Abstract
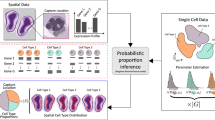
Spatial transcriptomic technologies promise to resolve cellular wiring diagrams of tissues in health and disease, but comprehensive mapping of cell types in situ remains a challenge. Here we present сell2location, a Bayesian model that can resolve fine-grained cell types in spatial transcriptomic data and create comprehensive cellular maps of diverse tissues. Cell2location accounts for technical sources of variation and borrows statistical strength across locations, thereby enabling the integration of single-cell and spatial transcriptomics with higher sensitivity and resolution than existing tools. We assessed cell2location in three different tissues and show improved mapping of fine-grained cell types. In the mouse brain, we discovered fine regional astrocyte subtypes across the thalamus and hypothalamus. In the human lymph node, we spatially mapped a rare pre-germinal center B cell population. In the human gut, we resolved fine immune cell populations in lymphoid follicles. Collectively, our results present сell2location as a versatile analysis tool for mapping tissue architectures in a comprehensive manner.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data generated for this manuscript (snRNA-seq and Visium from adjacent sections in the mouse brain (Fig. 2a), sequencing reads as well as Cell Ranger and Space Ranger output) were submitted to ArrayExpress under accession numbers E-MTAB-11114 (Visium) and E-MTAB-11115 (snRNA-seq). Annotated snRNA-seq data are publicly available via cellxgene portals64: full dataset (annotation_1_print column denotes cell types) and astrocyte subclusters. The integrated secondary lymphoid organ scRNA-seq data are publicly available for download through S3 bucket. Image data generated in this manuscript were deposited to the BioImage Archive (accession no. S-BIAD207).
Ground truth annotations of germinal center zones in lymph node Visium data can be found on GitHub. Ground truth annotations of the gut lymphoid follicles can be found on GitHub.
Published datasets. snRNA-seq data from Yao et al. were downloaded from the Allen Brain Institute data portal. Slide-seq V2 data from Stickels et al. were downloaded from the Broad Institute data portal (subject to use agreement). Visium data of human lymph nodes can be downloaded from the 10x website (via a function in the scanpy package).
Code availability
The cell2location package is available at https://github.com/BayraktarLab/cell2location/. Documentation and tutorials are available at https://cell2location.readthedocs.io/.
Code used to generate synthetic data is available at https://github.com/vitkl/cell2location_paper/blob/master/notebooks/benchmarking/synthetic_data_construction_improved_real_mg.ipynb. Code used to segment nuclei in histology images is available at https://github.com/yozhikoff/segmentation. Jupyter notebooks covering the analysis in this paper are available at https://github.com/vitkl/cell2location_paper/ and upon reasonable request.
References
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 3, 565 (2020).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00830-w (2021).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50 (2021).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Conde, C. D. et al. Cross-tissue immune cell analysis reveals tissue-specific adaptations and clonal architecture in humans. Preprint at https://www.researchgate.net/publication/351185031_Cross-tissue_immune_cell_analysis_reveals_tissue-specific_adaptations_and_clonal_architecture_across_the_human_body (2021).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353 (2018).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014 (2018).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 (2021).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Junker, J. P. et al. Genome-wide RNA tomography in the zebrafish embryo. Cell 159, 662–675 (2014).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
Aliee, H. & Theis, F. AutoGeneS: automatic gene selection using multi-objective optimization for RNA-seq deconvolution. Cell Syst. 12, 706–715 (2021).
Gayoso, A. et al. scvi-tools: a library for deep probabilistic analysis of single-cell omics data. Preprint at https://www.researchgate.net/publication/351290216_scvi-tools_a_library_for_deep_probabilistic_analysis_of_single-cell_omics_data (2021).
Lawson, C. L. & Hanson, R. J. Solving Least Squares Problems (SIAM, 1995).
Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016).
Phillips, J. W. et al. A repeated molecular architecture across thalamic pathways. Nat. Neurosci. 22, 1925–1935 (2019).
Lim, L., Mi, D., Llorca, A. & Marín, O. Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Ben Haim, L. & Rowitch, D. H. Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 18, 31–41 (2017).
Bayraktar, O. A. et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020).
Batiuk, M. Y. et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220 (2020).
Lanjakornsiripan, D. et al. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 9, 1623 (2018).
Chai, H. et al. Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95, 531–549 (2017).
Huang, A. Y.-S. et al. Region-specific transcriptional control of astrocyte function oversees local circuit activities. Neuron 106, 992–1008 (2020).
John Lin, C.-C. et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405 (2017).
Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020).
V1_Human_Lymph_Node - Datasets - Spatial Gene Expression - Official 10x Genomics Support. https://support.10xgenomics.com/spatial-gene-expression/datasets/1.1.0/V1_Human_Lymph_Node
James, K. R. et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353 (2020).
Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
King, H. W. et al. Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci. Immunol. 6, eabe6291 (2021).
Levitin, H. M. et al. De novo gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol. Syst. Biol. 15, e8557 (2018).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-seq. eLife 8, e43803 (2019).
Roco, J. A. et al. Class-switch recombination occurs infrequently in germinal centers. Immunity 51, 337–350 (2019).
Sun, B. T. Helper Cell Differentiation and Their Function (Springer, 2014).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
Fawkner-Corbett, D. et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 184, 810–826 (2021).
Spencer, J. & Sollid, L. M. The human intestinal B-cell response. Mucosal Immunol. 9, 1113–1124 (2016).
Koboziev, I., Karlsson, F. & Grisham, M. B. Gut-associated lymphoid tissue, T cell trafficking, and chronic intestinal inflammation. Ann. N. Y. Acad. Sci. 1207, E86–E93 (2010).
Cui, Y. et al. Astroglial Kir4.1 in the lateral habenula drives neuronal bursts in depression. Nature 554, 323–327 (2018).
Ward, L. M. The thalamus: gateway to the mind. Wiley Interdiscip. Rev. Cogn. Sci. 4, 609–622 (2013).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Roberts, K. et al. Transcriptome-wide spatial RNA profiling maps the cellular architecture of the developing human neocortex. Preprint at https://www.researchgate.net/publication/350237379_Transcriptome-wide_spatial_RNA_profiling_maps_the_cellular_architecture_of_the_developing_human_neocortex (2021).
Nichterwitz, S. et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 7, 12139 (2016).
Salvatier, J., Wiecki, T. V. & Fonnesbeck, C. Probabilistic programming in Python using PyMC3. PeerJ Comput. Sci. 2, e55 (2016).
Bingham, E. et al. Pyro: deep universal probabilistic programming. J. Mach. Learn. Res. 20, 973–978 (2019).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812 (2020).
Garcia-Alonso, L., Handfield, L. F. & Roberts, K. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 53, 1698–1711 (2021).
Vento-Tormo, R. et al. Single-cell roadmap of human gonadal development. Preprint at https://assets.researchsquare.com/files/rs-496470/v1_covered.pdf?c=1631867880 (2021).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Krishnaswami, S. R. et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499–524 (2016).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Roberts, K. & Bayraktar, O. A. Automation of multiplexed RNAscope single-molecule fluorescent in situ hybridization and immunohistochemistry for spatial tissue mapping. Methods Mol. Biol. 2148, 229–244 (2020).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Turner, J. S. et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586, 127–132 (2020).
Madissoon, E. et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 21, 1 (2019).
Megill, C. et al. chanzuckerberg/cellxgene: release 0.15.0. https://doi.org/10.5281/zenodo.3710410 (2020).
Acknowledgements
We thank B. Velten, Y. Huang and L. Marconato for feedback on the cell2location model; M. Prete and V. Kiselev for dockerizing the cell2location tool and creating the web portals for sharing our data; N. Kumasaka for helpful comments on single-cell analysis; S. Leonard and K. Polanski for help with spatial and single-nucleus data processing; K. James for advice on gut immune cell types; K. Roberts for advice on smFISH; J. E. Kwa for advice on snRNA-seq; J. Eliasova for illustrations and logo design; K. James advice on human gut immune cells; A. Antanaviciute, H. Koohy and A. Simmons for sharing gut Visium data; F. Obermeyer and M. Jankowiak for help with advanced use of pyro code base; and D. Rowitch and S. Teichmann for comments on the manuscript. H.W.K. was funded by a Sir Henry Wellcome Postdoctoral Fellowship (213555/Z/18/Z). This study was supported by Wellcome Trust Core Funding to O.A.B.
Author information
Authors and Affiliations
Contributions
V.K., O.S. and O.A.B. conceived the study. V.K. developed and implemented the cell2location model with feedback from O.S. and worked on validation, mouse brain, human lymph node, gut and benchmarking analyses. A.S. performed Slide-seq and hyperparameter sensitivity analysis, performed histology image segmentation, wrote data visualization modules and contributed to implementation of cell2location in pyro and benchmarking analyses. E.D. worked on generating synthetic data, mouse brain cell annotation and validation of cell type mapping and contributed to lymph node analysis. V.KE. and A.S. contributed to the development and interpretation of the model for estimating cell type signatures. A.L., M.G., A.A.I. and M.S.J. contributed to the developing of model architecture, writing the model and downstream analysis code, interpretation of the cell2location results as well as downstream clustering and NMF. A.A.R. performed benchmarking of stereoscope, supervised by R.V.-T. and O.S. A.G. contributed to implementation of cell2location using scvi-tools and pyro. H.W.K. contributed to lymph node data analysis and interpretation, supervised by L.J. R.E. contributed to gut data analysis. L.R. performed snRNA-seq experiments. L.T. performed Visium experiments. J.S.P. performed smFISH experiments and imaging. T.L. performed smFISH image processing and astrocyte segmentation analysis. V.K., O.S. and O.A.B. wrote the manuscript, with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biotechnology thanks Steven Sloan, Xiaoqun Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–27 and Supplementary Methods
Supplementary Files 1–13
Supplementary Files 1–13 show spatial cell and mRNA abundance (color) of all cell types (panels) estimated by cell2location in 10x Visium data for the respective tissues and samples.
Rights and permissions
About this article
Cite this article
Kleshchevnikov, V., Shmatko, A., Dann, E. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol 40, 661–671 (2022). https://doi.org/10.1038/s41587-021-01139-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-01139-4
This article is cited by
-
Spatial transcriptomic brain imaging reveals the effects of immunomodulation therapy on specific regional brain cells in a mouse dementia model
BMC Genomics (2024)
-
Distinct tumor-TAM interactions in IDH-stratified glioma microenvironments unveiled by single-cell and spatial transcriptomics
Acta Neuropathologica Communications (2024)
-
Spatiotemporal metabolomic approaches to the cancer-immunity panorama: a methodological perspective
Molecular Cancer (2024)
-
Advances in spatial transcriptomics and its applications in cancer research
Molecular Cancer (2024)
-
Single cell lineage tracing reveals clonal dynamics of anti-EGFR therapy resistance in triple negative breast cancer
Genome Medicine (2024)