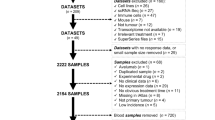
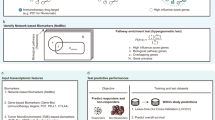
Abstract
Only a fraction of patients with cancer respond to immune checkpoint blockade (ICB) treatment, but current decision-making procedures have limited accuracy. In this study, we developed a machine learning model to predict ICB response by integrating genomic, molecular, demographic and clinical data from a comprehensively curated cohort (MSK-IMPACT) with 1,479 patients treated with ICB across 16 different cancer types. In a retrospective analysis, the model achieved high sensitivity and specificity in predicting clinical response to immunotherapy and predicted both overall survival and progression-free survival in the test data across different cancer types. Our model significantly outperformed predictions based on tumor mutational burden, which was recently approved by the U.S. Food and Drug Administration for this purpose1. Additionally, the model provides quantitative assessments of the model features that are most salient for the predictions. We anticipate that this approach will substantially improve clinical decision-making in immunotherapy and inform future interventions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All de-identified data needed to replicate all analyses are in Supplementary Table 3 and are available online at https://www.ioexplorer.org.
Code availability
The code used in this study is deposited at https://github.com/CCF-ChanLab/MSK-IMPACT-IO.
References
Subbiah, V., Solit, D. B., Chan, T. A. & Kurzrock, R. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥10: a decision centered on empowering patients and their physicians. Ann. Oncol. 31, 1115–1118 (2020).
Topalian, S. L., Taube, J. M., Anders, R. A. & Pardoll, D. M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 (2016).
Bendell, J. et al. Efficacy and safety results from IMblaze370, a randomised phase III study comparing atezolizumab plus cobimetinib and atezolizumab monotherapy vs regorafenib in chemotherapy-refractory metastatic colorectal cancer. Ann. Oncol. 29, 123–123 (2018).
Carbone, D. P. et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 376, 2415–2426 (2017).
Cohen, E. E. et al. Pembrolizumab (pembro) vs standard of care (SOC) for recurrent or metastatic head and neck squamous cell carcinoma (R/M HNSCC): phase 3 KEYNOTE-040 trial. Ann. Oncol. 28 https://oncologypro.esmo.org/meeting-resources/esmo-2017-congress/Pembrolizumab-pembro-vs-standard-of-care-SOC-for-recurrent-or-metastatic-head-and-neck-squamous-cell-carcinoma-R-M-HNSCC-Phase-3-KEYNOTE-040-trial (2017).
Powles, T. et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet 391, 748–757 (2018).
Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019).
Keenan, T. E., Burke, K. P. & Van Allen, E. M. Genomic correlates of response to immune checkpoint blockade. Nat. Med. 25, 389–402 (2019).
Anagnostou, V. et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat. Cancer 1, 99–111 (2020).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Rajkomar, A., Dean, J. & Kohane, I. Machine learning in medicine. N. Engl. J. Med. 380, 1347–1358 (2019).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumors: RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949 (2017).
Goodman, A. M. et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 (2017).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–20 (2019).
Luksza, M. et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017).
Valero, C. et al. The association between tumor mutational burden and prognosis is dependent on treatment context. Nat. Genet. 53, 11–15 (2021).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Chowell, D. et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 25, 1715–1720 (2019).
Chowell, D. et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018).
Mandal, R. et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364, 485–491 (2019).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Wang, Z. et al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat. Med. 25, 141–151 (2019).
Sanchez, A. et al. Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: a cohort study. Lancet Oncol. 21, 283–293 (2020).
Conforti, F. et al. Cancer immunotherapy efficacy and patients’ sex: a systematic review and meta-analysis. Lancet Oncol. 19, 737–746 (2018).
Jaillon, S. et al. Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer 20, 485–503 (2020).
Li, M. J. et al. Change in neutrophil to lymphocyte ratio during immunotherapy treatment is a non-linear predictor of patient outcomes in advanced cancers. J. Cancer Res. Clin. 145, 2541–2546 (2019).
Valero, C. et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat. Commun. 12, 729 (2021).
Kuai, J., Yang, F., Li, G. J., Fang, X. J. & Gao, B. Q. In vitro-activated tumor-specific T lymphocytes prolong the survival of patients with advanced gastric cancer: a retrospective cohort study. Onco Targets Ther. 9, 3763–3770 (2016).
Ikeguchi, A., Machiorlatti, M. & Vesely, S. K. Disparity in outcomes of melanoma adjuvant immunotherapy by demographic profile. Melanoma Manag 7, MMT43 (2020).
Jurasz, P., Alonso-Escolano, D. & Radomski, M. W. Platelet–cancer interactions: mechanisms and pharmacology of tumour cell-induced platelet aggregation. Br. J. Pharmacol. 143, 819–826 (2004).
Gupta, D. & Lis, C. G. Pretreatment serum albumin as a predictor of cancer survival: a systematic review of the epidemiological literature. Nutr. J. 9, 69 (2010).
Caro, J. J., Salas, M., Ward, A. & Goss, G. Anemia as an independent prognostic factor for survival in patients with cancer—a systematic, quantitative review. Cancer 91, 2214–2221 (2001).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Peng, D. et al. Prognostic significance of HALP (hemoglobin, albumin, lymphocyte and platelet) in patients with bladder cancer after radical cystectomy. Sci Rep. 8, 794 (2018).
Bindea, G., Mlecnik, B., Fridman, W. H., Pages, F. & Galon, J. Natural immunity to cancer in humans. Curr. Opin. Immunol. 22, 215–222 (2010).
Harrell, F. E. Jr., Lee, K. L. & Mark, D. B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15, 361–387 (1996).
Steyerberg, E. W. et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 21, 128–138 (2010).
Gurjao, C., Tsukrov, D., Imakaev, M., Luquette, L. J. & Mirny, L. A. Limited evidence of tumour mutational burden as a biomarker of response to immunotherapy. Preprint at https://www.biorxiv.org/content/10.1101/2020.09.03.260265v2 (2020).
Binnewies, M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018).
Krishna, C. et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell 39, 662–677 (2021).
Krishna, S. et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334 (2020).
Maier, B. et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262 (2020).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Krishna, C., Chowell, D., Gonen, M., Elhanati, Y. & Chan, T. A. Genetic and environmental determinants of human TCR repertoire diversity. Immun. Ageing 17, 26 (2020).
Braun, D. A. et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 26, 909–918 (2020).
Miao, D. et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 50, 1271–1281 (2018).
Patel, S. J. et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017).
Zaretsky, J. M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016).
Skoulidis, F. et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018).
Samstein, R. M. et al. Mutations in BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nat. Cancer 1, 1188–1203 (2020).
Wang, F. et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 5, 1504–1506 (2019).
Amin, M. B. et al. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging. CA Cancer J. Clin. 67, 93–99 (2017).
Zhou, J. et al. Analysis of tumor genomic pathway alterations using broad-panel next-generation sequencing in surgically resected lung adenocarcinoma. Clin. Cancer Res. 25, 7475–7484 (2019).
Shen, R. L. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Niu, B. F. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Shukla, S. A. et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 33, 1152–1158 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Pierini, F. & Lenz, T. L. Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol. Biol. Evol. 35, 2145–2158 (2018).
Robinson, J. et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–D431 (2015).
Zerbino, D. R. et al. Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018).
Grantham, R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Saito, T. & Rehmsmeier, M. Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics 33, 145–147 (2017).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.12, 77 (2011).
Schroder, M. S., Culhane, A. C., Quackenbush, J. & Haibe-Kains, B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 27, 3206–3208 (2011).
Mogensen, U. B., Ishwaran, H. & Gerds, T. A. Evaluating random forests for survival analysis using prediction error curves. J. Stat. Softw. 50, 1–23 (2012).
Acknowledgements
We thank the Chan lab and members of the Immunogenomics and Precision Oncology Platform for advice and input. This work was supported, in part, by NIH R35 CA232097 (T.A.C.), NIH RO1 CA205426 (T.A.C.), the PaineWebber Chair (T.A.C.), NIH/NCI Cancer Center Support Grant (P30 CA008748), Fundación Alfonso Martín Escudero (C.V.), NIH K08 DE024774, NIH R01 DE027738, the Sebastian Nativo Fund and the Jayme and Peter Flowers Fund (to L.G.T.M.).
Author information
Authors and Affiliations
Contributions
D.C., S.-K.Y., C.V., A.P., L.G.T.M., N.W. and T.A.C conceived and designed the study. D.C. and S.-K.Y. developed the machine learning model. D.C., S.-K.Y., C.V., A.P., C.K., M.L., D.H., H.S., D.W.K., N.P., V.M., K.W., T.L., R.M.S., N.R., P.S.A., V.P.B., G.P., A.A.H., A.N.S., M.A.P., R.J.M, M.L., A.Z., M.F.B., L.G.T.M. and N.W. acquired, analyzed or interpreted the data. M.G. provided statistical advice. All authors critically revised the manuscript for important intellectual content. L.G.T.M., N.W. and T.A.C. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
T.A.C. is a co-founder of Gritstone Oncology and holds equity. T.A.C. holds equity in An2H. T.A.C. acknowledges grant funding from Bristol Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H and Eisai. T.A.C. has served as an advisor for Bristol Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca and An2H. T.A.C., L.G.T.M. and D.C. hold ownership of intellectual property on using tumor mutational burden to predict immunotherapy response, with a pending patent, which has been licensed to PGDx. M.A.P. reports consulting fees from Bristol Myers Squibb, Merck, Array BioPharma, Novartis, Incyte, NewLink Genetics, Aduro and Eisai; honoraria from Bristol Myers Squibb and Merck; and institutional support from RGenix, Infinity, Bristol Myers Squibb, Merck, Array BioPharma, Novartis and AstraZeneca. M.L. has received advisory board compensation from Merck and Bristol Myers Squibb. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Biotechnology thanks Victor Velculescu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Supplementary Tables 1–3
Supplementary Table
Datasets
Rights and permissions
About this article
Cite this article
Chowell, D., Yoo, SK., Valero, C. et al. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat Biotechnol 40, 499–506 (2022). https://doi.org/10.1038/s41587-021-01070-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-01070-8
This article is cited by
-
Proteomic characterization of esophageal squamous cell carcinoma response to immunotherapy reveals potential therapeutic strategy and predictive biomarkers
Journal of Hematology & Oncology (2024)
-
Ultrasound-activated prodrug-loaded liposome for efficient cancer targeting therapy without chemotherapy-induced side effects
Journal of Nanobiotechnology (2024)
-
Exploiting tumor aneuploidy as a biomarker and therapeutic target in patients treated with immune checkpoint blockade
npj Precision Oncology (2024)
-
Identifying microRNAs associated with tumor immunotherapy response using an interpretable machine learning model
Scientific Reports (2024)
-
PIEZO1 mechanically regulates the antitumour cytotoxicity of T lymphocytes
Nature Biomedical Engineering (2024)