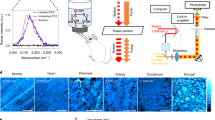
Abstract
We develop mid-infrared optoacoustic microscopy (MiROM) for label-free, bond-selective, live-cell metabolic imaging, enabling spatiotemporal monitoring of carbohydrates, lipids and proteins in cells and tissues. Using acoustic detection of optical absorption, MiROM converts mid-infrared sensing into a positive-contrast imaging modality with negligible photodamage and high sensitivity. We use MiROM to observe changes in intrinsic carbohydrate distribution from a diffusive spatial pattern to tight co-localization with lipid droplets during adipogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Cheng, J.-X. & Xie, X. S. Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science 350, aaa8870 (2015).
Baker, M. J. et al. Using fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 9, 1771–1791 (2014).
Diem, M. et al. Molecular pathology via IR and raman spectral imaging. J. Biophotonics 6, 855–886 (2013).
Prince, R. C., Frontiera, R. R. & Potma, E. O. Stimulated raman scattering: from bulk to nano. Chem. Rev. 117, 5070–5094 (2017).
Li, J. & Cheng, J.-X. Direct visualization of de novo lipogenesis in single living cells. Sci. Rep. 4, 6807 (2014).
Minamikawa, T. et al. Photo-induced cell damage analysis for single- and multifocus coherent anti-stokes raman scattering microscopy. J. Spectrosc. 2017, 1–8 (2017).
Yosef, H. K. et al. Noninvasive diagnosis of high-grade urothelial carcinoma in urine by raman spectral imaging. Anal. Chem. 89, 6893–6899 (2017).
Kallepitis, C. et al. Quantitative volumetric raman imaging of three dimensional cell cultures. Nat. Commun. 8, 14843 (2017).
Hu, F. et al. Vibrational imaging of glucose uptake activity in live cells and tissues by stimulated raman scattering. Angew. Chem. Int. Ed. Eng. 54, 9821–9825 (2015).
Chiu, L., Ho, S. H., Shimada, R., Ren, N. Q. & Ozawa, T. Rapid in vivo lipid/carbohydrate quantification of single microalgal cell by Raman spectral imaging to reveal salinity-induced starch-to-lipid shift. Biotechnol. Biofuels 10, 9 (2017).
Zhang, D. et al. Depth-resolved mid-infrared photothermal imaging of living cells and organisms with submicrometer spatial resolution. Sci. Adv. 2, e1600521 (2016).
Zhang, C. & Cheng, J. X. Perspective: coherent raman scattering microscopy, the future is bright. APL Photonics 3, 090901 (2018).
Haase, K., Kröger-Lui, N., Pucci, A., Schönhals, A. & Petrich, W. Real-time mid-infrared imaging of living microorganisms. J. Biophotonics 9, 61–66 (2016).
Martin, M. C. et al. 3D spectral imaging with synchrotron fourier transform infrared spectro-microtomography. Nat. Methods 10, 861–864 (2013).
Hong, W. et al. In situ detection of a single bacterium in complex environment by hyperspectral CARS imaging. ChemistrySelect 1, 513–517 (2016).
Liao, C.-S. et al. Stimulated raman spectroscopic imaging by microsecond delay-line tuning. Optica 3, 1377 (2016).
Oberg, K. A., Ruysschaert, J.-M. & Goormaghtigh, E. The optimization of protein secondary structure determination with infrared and circular dichroism spectra. Eur. J. Biochem 271, 2937–2948 (2004).
Lee, E. S. & Lee, J. Y. High resolution cellular imaging with nonlinear optical infrared microscopy. Opt. Express 19, 1378–1384 (2011).
Shi, J. et al. High-resolution, high-contrast mid-infrared imaging of fresh biological samples with ultraviolet-localized photoacoustic microscopy. Nat. Photonics 13, 609–615 (2019).
Aguirre, J. et al. Precision assessment of label-free psoriasis biomarkers with ultra-broadband optoacoustic mesoscopy. Nat. Biomed. Eng. 1, 0068 (2017).
Acknowledgements
The research leading to these results has received funding from the Deutsche Forschungsgemeinschaft (DFG), Germany (Gottfried Wilhelm Leibniz Prize 2013; NT 3/10-1), as well as from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program under grant agreement No 694968 (PREMSOT). The authors thank A. Georgiadi for advice, discussion and support on mouse adipocyte tissue preparation.
Author information
Authors and Affiliations
Contributions
M.A.P. created the imaging concept, designed, built and characterized the imaging system. M.A.P., A.A.K. and J.R. designed and performed the experiments on adipocytes. M.A.P. and J.R. designed and performed the experiments on excised tissues. A.C. synchronized and automated the imaging system. M.R.S. performed the image processing and prepared the art work. M.A.P. and A.S. designed and performed the molecular contrast validation experiments and viability tests on cells. M.A.P., F.G. and B.S. designed and performed the spectral validation of the system. M.A.P. processed the results, prepared the images, and wrote the manuscript. M.A.P., A.A., J.R. and M.S. analyzed the results on lipolysis. M.S. and S.H. supervised the study on lipolysis. V.N. supervised the whole study. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
V.N. has financial interests in iThera Medical (equity owner and consultant); SurgVision B /Bracco Sp.A (member of the scientific advisory board); and Spear UG (owner). These companies, however, did not contribute to this work. V.N. and M.A.P. are inventors on a provisional patent application related to mid-infrared optoacoustic microscopy.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Limit of detection of MiROM for DMSO and albumin.
(a) Normalized optoacoustic spectra (NOAS) of water solutions of DMSO at different concentrations (after water subtraction). The absorption band due to the S=O vibration of the DMSO molecule is clearly recognized above the baseline (BL) down to a concentration of 2.5 mM. (b) Area of NOAS for the S=O absorption band vs concentration of DMSO, the lowest detected concentration with a SNR > 2 is indicates by the red arrow (2.5 mM with a SNR of 6). (c) MiROM micrograph of D2O and carbon tape at 1650 cm−1. The LOD for albumin was tested changing its concentration from 0 to 757 µM while measuring the OA signal at the indicated point S. The OA signal at point R in the carbon tape served as spectral reference. (d) NOAS of D2O solutions of albumin at different concentrations (after D2O subtraction). The amide I absorption band of albumin is clearly recognized above the BL down to a concentration of 1.5 µM. (e) Area of NOAS of the amide I band vs concentration of albumin, the lowest detected concentration with a SNR > 2 is indicates by the red arrow (1.5 µM with a SNR of 4.5). (f) Contrast test of albumin solutions in the area indicated in c (red dashed line) showing that: images of albumin with concentrations as low as 1.5 µM can be obtained and recognized above the contrast level of D2O. (g) Contrast test of the carbon tape (for reference) in the area indicated in c (black dashed line). Data in a-g are representative of three independent experiments. In b,e the error bars represent: area of NOAS ± noise level (see Methods).
Supplementary Fig. 2 Cell viability test on 3T3-L1 cells upon mid-infrared laser irradiation.
Each bar represents the mean value of cell viability of control (CNT) and irradiated (IRR) samples obtained from three independent experiments (n=3). The error bars represent the standard deviation (SD) of the cell viability. By unpaired t-test, a two-tailed P = 0.77 was obtained, meaning that the hypothesized contribution of an increased cell death induced by laser exposure can be rejected.
Supplementary Fig. 3 Endogenous contrast coverage and spectral validation of MiROM.
(a,b) Comparison of mid-IR absorption spectrum of selected biomolecules measured by standard ATR-FTIR spectroscopy (blue lines) and MiROM (red lines). (a) DNA and (b) albumin in D2O. The vertical dashed lines indicate a good matching of the main absorption bands measured by standard ATR-FTIR and mid-IR optoacoustic spectroscopy. A detailed comparison of the position of the measured absorption bands for each biomolecule is shown in Supplementary Tables 2–5. (c-e) MiROM imaging/spectroscopy of water. (c) Micrograph of water and carbon tape at 1650 cm−1. Arrow 1 indicates where the optoacoustic signal of water was recorded while arrow 2 indicates where the reference signal was measured from the carbon tape, scale bar: 200 μm. (d) MiROM spectrum of water. The dashed line indicates the wavelength (1650 cm-1) used to obtain the image in c. (e) Intensity profile at the white-dashed line in c showing the contrast level in water, carbon tape, and voids in the carbon tape. Data in a-e are representative of three independent experiments.
Supplementary Fig. 4 Description of the imaging system, spatial resolution and contrast test.
(a) Schematic diagram of the imaging system. A tunable pulsed Quantum Cascade Laser (QCL) provides the excitation for optoacoustic imaging. An absorption intensity map is obtained by displacing the sample along the focal plane while simultaneously acquiring the optoacoustic signal. (b,c) Lateral and axial resolution test of the imaging system. (b, upper) Point spread function (PSF) using a 1 µm polystyrene sphere. (b, lower) Intensity profile along different orientations of the PSF. The mean full-width at the half maximum (FWHM) indicates a lateral resolution of 5.3 µm. (c, upper) Intensity projection in z of the same PSF shown in b. (c, lower) Intensity profile along a selected line in the z projection. The FWHM indicates an axial resolution of 42.2 µm. An acoustic reflection from the ZnSe window of the sample holder (FWHM of 58.1 µm) overlapping with the main signal was also observed. (d) Contrast test of the endogenous imaging system using a 10 µm polyamide suture and 50 µm polyethylene spheres; FOV 0.5 mm x 1.0 mm and 2 µm step size. (left) Image obtained at 2850 cm-1 showing a strong positive contrast for the spheres and a dim positive contrast for the suture; the background (agar) appears dark. (right) Image obtained at 1550 cm-1. Here the strong contrast comes from the suture, but also the water content in agar offers positive contrast. In this image, the polyethylene spheres appear dark, scale bars: 100 µm. Data in b-d are representative of three independent experiments.
Supplementary Fig. 5 Imaging cell membrane in undifferentiated preadipocytes and HeLa cells.
MiROM micrographs at 2853 cm-1 of (a) PreBAT, (b) 3T3-L1, and (c) HeLa cells after background subtraction. The FOV for a and b is 125 µm x 125 µm while the FOV for c is 150 µm x 150 µm, scale bars: 25 µm. The contrast profile at the indicated dashed line on each cell is plotted below the regarding micrograph. The CNR in PreBAT is 22:1, in 3T3-L1 is 17:1, and in HeLa cells is 7:1. The units in the contrast bars are in mV. Images a-c are representative of three independent experiments.
Supplementary Fig. 6 Intrinsic contrast profiles in living cells and tissues.
(a,b) Contrast test of the 3T3-L1 adipocytes and pancreatic mouse tissue, respectively, in main Fig. 1, scale bars: 100 µm. The units in the contrast bars are mV. The contrast profile of each cell at the indicated dashed lines (blue for lipids and green for proteins) is plotted in the lower part of a. Data in a,b are representative of five independent experiments.
Supplementary Fig. 7 Comparison between MiROM carbohydrate contrast and total carbohydrate colorimetric assay absorptivity.
In (a) 3T3-L1 cells cultured in differentiation media (high glucose, 25 mM) until day 6 and in growth media (low glucose, 5 mM) afterwards. (b,c) 3T3-L1 cells cultured in differentiation media until day 6, on which cells are incubated for 48h (d8) using three different culturing conditions: in growth media (Low gluc, 5 mM), in differentiation media (High gluc, 25 mM) and in differentiation media with 5 nM of GLUT1 inhibitor (GLUT1 Inh). In both, a and b, the mean CBR/c for micrographs of differentiating 3T3-L1 cells measured at 1022 cm-1 fairly correlates with the read-outs obtained by the total carbohydrate assay applied to the imaged dishes (see Methods). Carbohydrate contrast obtained by MiROM accurately reflects the glucose stimulation of the imaged cells accordingly. In a, for both methods each measurement point is the average of two cell-dishes; including the measurement series shown in main Fig. 2f–n. Only the measurement points at day 10 in a slightly shifted from the same trend, most probably reflecting the presence of additional molecules detectable by MiROM for which the assay is not sensitive, e.g. glycerol with an absorption band around 1022 cm-1. Data in a,c are presented as dot plots (n=2 per time point; error bars represent mean ± s.d.).
Supplementary Fig. 8 Co-localization of lipid and carbohydrate contrast in adipocytes.
At incubation day 6 (a), the carbohydrate contrast is broadly spread around LDs formation, while at incubation day 14 (b) it is highly co-localized with mature LDs. The Pearson Correlation Coefficient (PCC) between channels in c and d confirms this observation, n=90000. Scale bars: 200 µm. Images a,b are representative of three independent experiments.
Supplementary Fig. 9 Label-free monitoring of lipids and proteins in adipocytes.
(a) Monitoring of induced lipolysis in brown (differentiated PreBAT) adipocytes; lipid maps at 2857 cm-1. Two ROIs enclosing individual adipocytes are marked on each sequence, green dashed circle for ROI 1 and red dashed circle for ROI 2. Time and presence of ISO is indicated at the bottom corners of each frame. Overall relative lipid and protein contrast change for (b) white and (c) brown adipocytes. (d) Relative lipid contrast change for ROIs 1 and 2 of the brown adipocytes in a. The red arrow in b-d indicates when ISO was added. (e) Complementary merged lipid (yellow)-protein (blue) map and protein-only map (grey) of cells in main Fig. 1 m, n. (f) Complementary merged lipid (yellow)-protein (blue) map and protein-only map (grey) of cells in a, scale bars: 50 µm. See also Supplementary Videos 1a, 1b and 2a, 2b. Data in a,c,d,f are representative of three independent experiments. Data in b,e are representative of five independent experiments.
Supplementary Fig. 10 Label-free monitoring of protein dynamics during lipolysis.
(a) Protein map (at 1550 cm-1) of white adipocytes. Here, three ROIs have been marked: blue dashed square for ROI 1, black dashed square for ROI 2, and green dashed square for ROI 3. Each ROI is centered on a single adipocyte. (b-d) Monitoring of proteins during lipolysis for the selected ROIs in g. The white arrow follows regions changing protein contrast during lipolysis. Common time and presence of ISO for each ROI, is indicated above sequence b. The FOV of a is 250 µm x 250 µm. See also Supplementary Videos 3a, 3b. (e,f) Complementary merged lipid (yellow)-protein (blue) map and lipid-only map (grey) of the cells in a-d. Images a-f are representative of three independent experiments.
Supplementary Fig. 11 MiROM micrographs of thick frozen tissue sections.
(a) Brightfield image of a transverse whole-body cryosection of a mouse; the lungs and the heart ventricles are clearly identified. (b) Merged lipid (at 2850 cm-1, yellow) and protein (at 1550 cm-1, blue) map produced by MiROM of the same section in a, indicating two ROIs, total imaging time 26 min. (c) (left) Protein-map, (center) lipid-map, and (right) merged lipid/protein map of the zoomed ROI 1, scale bar: 200 µm, total imaging time 54 min. (d) (left) Protein-map, (center) lipid-map, and (right) merged lipid/protein image of the zoomed ROI 2, scale bar: 250 µm, total imaging time 4 min. ROI 1 shows a lipid-containing deposit within a groove-like space of relatively homogeneous protein contrast, most likely part of the lymphatic vascular network. ROI 2 shows sparsely distributed lipid droplets in the left ventricular cardiac muscle. Contrast bars in mV. Images a-d are representative of five independent experiments.
Supplementary Fig. 12 Large MiROM micrographs of freshly excised muscle and fat tissue.
(a) Gastrocnemius muscle (GC muscle). (b) Inguinal White Adipocyte Tissue (iWAT). (c) Inguinal Brown Adipocyte Tissue (iBAT). The high-contrast MiROM micrographs reveal the heterogenous distribution of lipids (at 2850 cm-1), proteins (at 1650 cm-1), and carbohydrates (at 1022 cm-1) in fresh unprocessed tissue samples. Imaging time ~20 min per contrast channel for the large FOVs and 9 min per contrast channel for the zoom in c. Images a-c are representative of four independent experiments.
Supplementary Fig. 13 Imaging depth of the pancreatic tissue in main Fig. 1.
(a) Depth map of the acinar glands detected at 2850 cm-1 calculated using the time arrival of OA signals, depth is color-coded according the contrast bar to the left. Two points (P1 and P2) from two different structures at different depths are indicated. (b) OA intensity map of a showing also P1 and P2 for comparison. (c) 3D surface plot of the depth map in a. (d) Comparison of the time arrival of the OA signal detected at P1 and P2. Data in a-d are representative of three independent experiments.
Supplementary Fig. 14 Imaging depth characterization of MiROM for a polyamide suture embedded in pure fat (lard).
(a) Brightfield image of the phantom array showing two sutures embedded in fat in a measurement cuvette (only one suture was measured). One end of the sutures is attached to the surface of the cuvette while the other end is held by a spacer above the cuvette’s surface, creating an elevation angle of the sutures regarding the surface. (b) Comparison of time arrivals of the OA (PA) signal (envelope) at different positions along the suture’s length, the positions are indicated in c by dashed lines. The legend shows the calculated depth from which the signals arrive. (c) Top view (x-y) maximum amplitude projection of the OA signal, the colored dashed lines correspond to the envelope plots in b. (d) Side view (y-z) maximum amplitude projection of the OA signal. Here, the depth of the OA signal with a SNR of at least 2.4 (~7.6 dB) is indicated. This was found at ~575 µm (L2 in b). Data in a-d are representative of two independent experiments.
Supplementary Fig. 15 Comparison between MiROM and Laser Scanning Confocal Microscopy (LSCM) micrographs of lipid droplets.
Fixed 3T3-L1 adipocytes (incubation day 8) imaged by (a) MiROM and (b) LSCM upon Nile red staining, FOV 150 µm x 150 µm. Both modalities reveal a co-localized pattern (PCC: 0.84, n=22,500) indicating the congruent information content. Missing information in the MiROM image can be explained by the significant lower spatial resolution achieved in the mid-IR range at 3500 nm (2850 cm-1) than the VIS excitation wavelength for confocal microscopy at 510 nm. Several small lipid droplets or lipid droplet clusters observed in the confocal microscopy image, are not resolved as individual structures in MiROM. They are, however, still detected as rather homogenous contrast distribution. Furthermore, in the MiROM image, the cell bodies can be observed due to their lipid content in the cell membrane, whereas the LSCM lacks in revealing the cell membrane.
Supplementary information
Supplementary Materials
Supplementary Figs. 1–15 and Supplementary Tables 1–6.
Supplementary Video 1
Lipid contrast monitoring before and after induced lipolysis on PreBAT adipocytes. The average contrast of two regions of interest, each one enclosing a single cell, have been selected for comparison of the lipolysis rate in these two cells. Data are representative of three independent experiments.
Supplementary Video 2
Lipid and protein contrast monitoring before and after induced lipolysis on PreBAT adipocytes.The overall change of lipid and protein contrast of the field of view is plotted to remark the different behavior between lipids and proteins in the cells. Data are representative of three independent experiments.
Supplementary Video 3
Lipid contrast monitoring before and after induced lipolysis on 3T3-L1 adipocytes. The average contrast of two regions of interest, each one enclosing a single cell, have been selected for comparison of the lipolysis rate in these two cells. Data representative of five independent experiments.
Supplementary Video 4
Lipid and protein contrast monitoring before and after induced lipolysis on 3T3-L1 adipocytes. The overall change of lipid and protein contrast of the field of view is plotted to remark the different behavior between lipids and proteins in the cells. Data are representative of five independent experiments.
Supplementary Video 5
Protein contrast monitoring before and after induced lipolysis on 3T3-L1 adipocytes. Three regions of interest, each one enclosing a single cell, have been zoomed to highlight the dynamic behavior of proteins during lipolysis. Data are representative of three independent experiments.
Supplementary Video 6
Protein and lipid contrast monitoring before and after induced lipolysis on 3T3-L1 adipocytes. Three regions of interest, each one enclosing a single cell, have been zoomed to highlight the dynamic behavior of proteins and lipids during lipolysis. Data representative of three independent experiments.
Rights and permissions
About this article
Cite this article
Pleitez, M.A., Khan, A.A., Soldà, A. et al. Label-free metabolic imaging by mid-infrared optoacoustic microscopy in living cells. Nat Biotechnol 38, 293–296 (2020). https://doi.org/10.1038/s41587-019-0359-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0359-9
This article is cited by
-
Non-invasive measurements of blood glucose levels by time-gating mid-infrared optoacoustic signals
Nature Metabolism (2024)
-
Blood glucose concentration measurement without finger pricking
Nature Metabolism (2024)
-
An engineering makeover of biomedical research
Nature Reviews Bioengineering (2023)
-
Bond-selective fluorescence imaging with single-molecule sensitivity
Nature Photonics (2023)
-
Infrared spectroscopic laser scanning confocal microscopy for whole-slide chemical imaging
Nature Communications (2023)