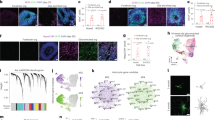
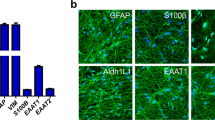
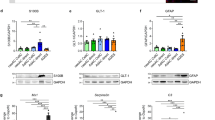
Abstract
The mechanistic basis of gliogenesis, which occurs late in human development, is poorly understood. Here we identify nuclear factor IA (NFIA) as a molecular switch inducing human glial competency. Transient expression of NFIA is sufficient to trigger glial competency of human pluripotent stem cell-derived neural stem cells within 5 days and to convert these cells into astrocytes in the presence of glial-promoting factors, as compared to 3–6 months using current protocols. NFIA-induced astrocytes promote synaptogenesis, exhibit neuroprotective properties, display calcium transients in response to appropriate stimuli and engraft in the adult mouse brain. Differentiation involves rapid but reversible chromatin remodeling, glial fibrillary acidic protein (GFAP) promoter demethylation and a striking lengthening of the G1 cell cycle phase. Genetic or pharmacological manipulation of G1 length partially mimics NFIA function. We used the approach to generate astrocytes with region-specific or reactive features. Our study defines key mechanisms of the gliogenic switch and enables the rapid production of human astrocytes for disease modeling and regenerative medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data and reagents in this study are available from the corresponding author upon reasonable request. All FASTQ files and Supplementary files were uploaded to National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE104232.
References
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Molofsky, A. V. et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 26, 891–907 (2012).
Sauvageot, C. M. & Stiles, C. D. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 12, 244–249 (2002).
Studer, L., Vera, E. & Cornacchia, D. Programming and reprogramming cellular age in the era of induced pluripotency. Cell Stem Cell 16, 591–600 (2015).
Tcw, J. et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Reports 9, 600–614 (2017).
Krencik, R. et al. Dysregulation of astrocyte extracellular signaling in Costello syndrome. Sci. Transl. Med. 7, 286ra266 (2015).
Tao, Y. & Zhang, S. C. Neural subtype specification from human pluripotent stem cells. Cell Stem Cell 19, 573–586 (2016).
Chandrasekaran, A., Avci, H. X., Leist, M., Kobolak, J. & Dinnyes, A. Astrocyte differentiation of human pluripotent stem cells: new tools for neurological disorder research. Front. Cell. Neurosci. 10, 215 (2016).
Santos, R. et al. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Reports 8, 1757–1769 (2017).
Krencik, R., Weick, J. P., Liu, Y., Zhang, Z. J. & Zhang, S. C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 29, 528–534 (2011).
Stolt, C. C. et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 (2003).
Deneen, B. et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006).
Patterson, M. et al. let-7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Reports 3, 758–773 (2014).
Naka, H., Nakamura, S., Shimazaki, T. & Okano, H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat. Neurosci. 11, 1014–1023 (2008).
Hirabayashi, Y. et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613 (2009).
Koch, P., Opitz, T., Steinbeck, J. A., Ladewig, J. & Brustle, O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl Acad. Sci. USA 106, 3225–3230 (2009).
Elkabetz, Y. et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22, 152–165 (2008).
Liu, Y. et al. CD44 expression identifies astrocyte-restricted precursor cells. Dev. Biol. 276, 31–46 (2004).
Chen, H. et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 14, 796–809 (2014).
Calder, E. L. et al. Retinoic acid-mediated regulation of GLI3 enables efficient motoneuron derivation from human ESCs in the absence of extrinsic SHH activation. J. Neurosci. 35, 11462–11481 (2015).
Magistri, M. et al. A comparative transcriptomic analysis of astrocytes differentiation from human neural progenitor cells. Eur. J. Neurosci. 44, 2858–2870 (2016).
Lovatt, D. et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J. Neurosci. 27, 12255–12266 (2007).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Ullian, E. M., Sapperstein, S. K., Christopherson, K. S. & Barres, B. A. Control of synapse number by glia. Science 291, 657–661 (2001).
Sofroniew, M. V. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16, 249–263 (2015).
Allen, N. J. & Eroglu, C. Cell biology of astrocyte-synapse interactions. Neuron 96, 697–708 (2017).
Betz, A. et al. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron 21, 123–136 (1998).
Sofroniew, M. V. & Vinters, H. V. Astrocytes: biology and pathology. Acta Neuropathol. 119, 7–35 (2010).
Liddelow, S. A. & Barres, B. A. Reactive astrocytes: production, function, and therapeutic potential. Immunity 46, 957–967 (2017).
Cornell-Bell, A. H., Finkbeiner, S. M., Cooper, M. S. & Smith, S. J. Glutamate induces calcium waves in cultured astrocytes: long-range glial signaling. Science 247, 470–473 (1990).
Oberheim, N. A. et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 29, 3276–3287 (2009).
Rajan, P. & McKay, R. D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 18, 3620–3629 (1998).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Takahashi, T., Nowakowski, R. S. & Caviness, V. S. Jr. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 15, 6046–6057 (1995).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Calegari, F. & Huttner, W. B. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J. Cell. Sci. 116, 4947–4955 (2003).
Vodermaier, H. C. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14, R787–R796 (2004).
Sigl, R. et al. Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J. Cell. Sci. 122, 4208–4217 (2009).
Garcia-Campmany, L. & Marti, E. The TGFbeta intracellular effector Smad3 regulates neuronal differentiation and cell fate specification in the developing spinal cord. Development 134, 65–75 (2007).
Zhang, Y., Alexander, P. B. & Wang, X. F. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol 9, a022145 (2017).
Zeltner, N. & Studer, L. Pluripotent stem cell-based disease modeling: current hurdles and future promise. Curr. Opin. Cell Biol. 37, 102–110 (2015).
Sances, S. et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 19, 542–553 (2016).
Williams, E. C. et al. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum. Mol. Genet. 23, 2968–2980 (2014).
Le Roux, P. D. & Reh, T. A. Astroglia demonstrate regional differences in their ability to maintain primary dendritic outgrowth from mouse cortical neurons in vitro. J. Neurobiol. 27, 97–112 (1995).
Holmqvist, S. et al. Generation of human pluripotent stem cell reporter lines for the isolation of and reporting on astrocytes generated from ventral midbrain and ventral spinal cord neural progenitors. Stem Cell Res. 15, 203–220 (2015).
das Neves, L. et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl Acad. Sci. USA 96, 11946–11951 (1999).
Steele-Perkins, G. et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 25, 685–698 (2005).
Canals, I. et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Methods 15, 693–696 (2018).
Pauklin, S. & Vallier, L. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013).
Tchieu, J. et al. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 21, 399–410 e397 (2017).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Steinbeck, J. A. et al. Functional connectivity under optogenetic control allows modeling of human neuromuscular disease. Cell Stem Cell 18, 134–143 (2016).
Chiricozzi, E. et al. Group IIA secretory phospholipase A2 (GIIA) mediates apoptotic death during NMDA receptor activation in rat primary cortical neurons. J. Neurochem. 112, 1574–1583 (2010).
Cheng, P. Y. et al. Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS ONE 6, e22018 (2011).
Maroof, A. M. et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 12, 559–572 (2013).
Ying, S. W. & Goldstein, P. A. Propofol suppresses synaptic responsiveness of somatosensory relay neurons to excitatory input by potentiating GABA(A) receptor chloride channels. Mol. Pain 1, 2 (2005).
Acknowledgements
We are grateful to the members the Studer laboratory for helpful discussions and support for this project, and to G. Cederquist, M. Tomishima, S. Irion and V. Tabar for their critical comments on the manuscript. We would also like to give special thanks to A. Koff (MSKCC) for his helpful comments, experimental discussions regarding the cell cycle and comments on the manuscript. Additionally, we would like to thank A. Viale at Integrated Genomics Operation Core (MSKCC) for the RNA-sequencing studies, M. Witkin at the Epigenetics Core (MSKCC) for the ATAC sequencing, S. Fujisawa, E. Feng and V. Boyko at the Molecular Cytology Core (MSKCC) for help in calcium imaging studies and quantification of synaptic proteins, R. Garripa and H. Liu at the RNAi Core (MSKCC) for help with short hairpin RNA design and the Flow Cytometry Core (MSKCC) for the cell-sorting applications. J.T. was supported by the Tri-I Starr Stem Cell Scholars postdoctoral training fellowship. S.R.G was supported by the Ruth L. Kirschstein Individual Predoctoral NRSA for MD/PhD Fellowship (No. 1F30MH115616-01) and by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health (No. T32GM007739) to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. E.M.G. was supported by a grant from the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (No. 323630-164217). The work was supported by grants to L.S. from the National Institutes of Health (No. R21NS084334, No. R01AG056298), by core grant No. P30CA008748 and by a grant from Project ALS.
Author information
Authors and Affiliations
Contributions
J.T. contributed to the conception, study design, data analysis and interpretation, writing of the manuscript, bioinformatics, development and execution of directed differentiation strategies from hPSCs, generation of LTNSCs, cell cycle analysis and calcium imaging. E.L.C. contributed to maintenance of hPSCs and directed differentiation of spinal cord progenitors. S.R.G. contributed to cell cycle analysis and astrocyte activation assays. E.M.G.contributed to transplantation studies and data analysis. K.A.A. and P.A.G. contributed to electrophysiology and assessment of neuronal maturation. J.A.S. contributed to generation of LTNSCs and calcium imaging. L.S. contributed to the conception, study design, data analysis and interpretation and writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The Memorial Sloan-Kettering Cancer Center has filed a patent application (WO2018175574A1) on the methods described in the manuscript. L.S. is the scientific cofounder of Bluerock Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–25
Supplementary Table 1
Gene expression dataset of NFIA-induced astrocytes log transformed counts per million (CPM)
Supplementary Table 2
Gene expression dataset of the NFIA-induction time course. Fold changes compared to day 0 (neurogenic NSCs)
Supplementary Table 3
List of antibodies used in this study
Supplementary Video 1
FUCCI-O reporter on LTNSCs without doxycycline induction
Supplementary Video 2
FUCCI-O reporter on LTNSCs with continuous doxycycline treatment
Rights and permissions
About this article
Cite this article
Tchieu, J., Calder, E.L., Guttikonda, S.R. et al. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37, 267–275 (2019). https://doi.org/10.1038/s41587-019-0035-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0035-0
This article is cited by
-
Mutual interaction of neurons and astrocytes derived from iPSCs with APP V717L mutation developed the astrocytic phenotypes of Alzheimer’s disease
Inflammation and Regeneration (2024)
-
Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain
Nature Biotechnology (2024)
-
Gliomas: a reflection of temporal gliogenic principles
Communications Biology (2024)
-
Combined small-molecule treatment accelerates maturation of human pluripotent stem cell-derived neurons
Nature Biotechnology (2024)
-
Gatad2b, associated with the neurodevelopmental syndrome GAND, plays a critical role in neurodevelopment and cortical patterning
Translational Psychiatry (2024)