Abstract
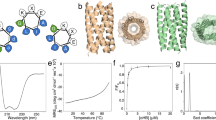
The α-helix is pre-eminent in structural biology1 and widely exploited in protein folding2, design3 and engineering4. Although other helical peptide conformations do exist near to the α-helical region of conformational space—namely, 310-helices and π-helices5—these occur much less frequently in protein structures. Less favourable internal energies and reduced tendencies to pack into higher-order structures mean that 310-helices rarely exceed six residues in length in natural proteins, and that they tend not to form normal supersecondary, tertiary or quaternary interactions. Here we show that despite their absence in nature, synthetic peptide assemblies can be built from 310-helices. We report the rational design, solution-phase characterization and an X-ray crystal structure for water-soluble bundles of 310-helices with consolidated hydrophobic cores. The design uses six-residue repeats informed by analysing 310-helical conformations in known protein structures, and incorporates α-aminoisobutyric acid residues. Design iterations reveal a tipping point between α-helical and 310-helical folding, and identify features required for stabilizing assemblies of 310-helices. This work provides principles and rules to open opportunities for designing into this hitherto unexplored region of protein-structure space.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The coordinate and structure factor files are available from the Research Collaboratory for Structural Bioinformatics PDB under the following accession codes: CCTri-TypeN-LaLd (PDB ID: 7QDK); D-310HD (PDB ID: 7QDI); PK-10 + PK-11 (PDB ID: 7QDJ). The list of PDB files for the bioinformatic analyses was downloaded from the Pisces server (http://dunbrack.fccc.edu/pisces/download/). Source data are provided with this paper. Additional data to generate figures in the Supplementary Information are available at http://coiledcoils.chm.bris.ac.uk/SI-data/PK-310/.
Code availability
The customized scripts used for bioinformatic analyses are available at http://coiledcoils.chm.bris.ac.uk/SI-data/PK-310/.
References
Pauling, L., Corey, R. B. & Branson, H. R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl Acad. Sci. USA 37, 205–211 (1951).
Chakrabartty, A. & Baldwin, R. L. Stability of alpha-helices. Adv. Protein Chem. 46, 141–176 (1995).
Korendovych, I. V. & DeGrado, W. F. De novo protein design, a retrospective. Q. Rev. Biophys. 53, e3 (2020).
Lapenta, F., Aupic, J., Strmsek, Z. & Jerala, R. Coiled coil protein origami: from modular design principles towards biotechnological applications. Chem. Soc. Rev. 47, 3530–3542 (2018).
Schulz, G. E. & Schirmer, R. H. Principles of Protein Structure (Springer, 1979).
Scholtz, J. M. & Baldwin, R. L. The mechanism of alpha-helix formation by peptides. Annu. Rev. Biophys. Biomol. Struct. 21, 95–118 (1992).
Woolfson, D. N. A brief history of de novo protein design: minimal, rational, and computational. J. Mol. Biol. 433, 167160 (2021).
Dawson, W. M., Rhys, G. G. & Woolfson, D. N. Towards functional de novo designed proteins. Curr. Opin. Chem. Biol. 52, 102–111 (2019).
Ramachandran, G. N., Ramakrishnan, C. & Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7, 95–99 (1963).
Kuster, D. J., Liu, C., Fang, Z., Ponder, J. W. & Marshall, G. R. High-resolution crystal structures of protein helices reconciled with three-centered hydrogen bonds and multipole electrostatics. PLoS ONE 10, e0123146 (2015).
Burley, S. K. et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 49, D437–D451 (2021).
Chothia, C., Levitt, M. & Richardson, D. Helix to helix packing in proteins. J. Mol. Biol. 145, 215–250 (1981).
Orengo, C. A. et al. CATH–a hierarchic classification of protein domain structures. Structure 5, 1093–1108 (1997).
Gessmann, R., Axford, D., Owen, R. L., Bruckner, H. & Petratos, K. Four complete turns of a curved 310-helix at atomic resolution: the crystal structure of the peptaibol trichovirin I-4A in a polar environment suggests a transition to α-helix for membrane function. Acta Crystallogr. D 68, 109–116 (2012).
Toniolo, C. & Brückner, H. Peptaibiotics (Wiley, 2009).
Toniolo, C. & Benedetti, E. The polypeptide 310-helix. Trends Biochem. Sci. 16, 350–353 (1991).
Gessmann, R., Bruckner, H. & Petratos, K. The crystal structure of Z-(Aib)10-OH at 0.65 Å resolution: three complete turns of 310-helix. J. Pept. Sci. 22, 76–81 (2016).
Solà, J., Helliwell, M. & Clayden, J. Interruption of a 310-helix by a single Gly residue in a poly-Aib motif: a crystallographic study. Biopolymers 95, 62–69 (2011).
Pike, S. J., Boddaert, T., Raftery, J., Webb, S. J. & Clayden, J. Participation of non-aminoisobutyric acid (Aib) residues in the 310 helical conformation of Aib-rich foldamers: a solid state study. New J. Chem. 39, 3288–3294 (2015).
Karle, I. L., Flippen-Anderson, J. L., Gurunath, R. & Balaram, P. Facile transition between 310- and α-helix: structures of 8-, 9-, and 10-residue peptides containing the -(Leu-Aib-Ala)2-Phe-Aib-fragment. Protein Sci. 3, 1547–1555 (1994).
Toniolo, C. et al. Preferred conformation of the terminally blocked (Aib)10 homo-oligopeptide: a long, regular 310-helix. Biopolymers 31, 129–138 (1991).
Nagaraj, R. & Balaram, P. Alamethicin, a transmembrane channel. Acc. Chem. Res. 14, 356–362 (1981).
Toniolo, C. et al. Conformation of pleionomers of .alpha.-aminoisobutyric acid. Macromolecules 18, 895–902 (1985).
Karle, I. L. & Balaram, P. Structural characteristics of alpha-helical peptide molecules containing Aib residues. Biochemistry 29, 6747–6756 (1990).
Byrne, L. et al. Foldamer-mediated remote stereocontrol: >1,60 asymmetric induction. Angew. Chem. Int. Ed. 53, 151–155 (2014).
Lister, F. G. A., Le Bailly, B. A. F., Webb, S. J. & Clayden, J. Ligand-modulated conformational switching in a fully synthetic membrane-bound receptor. Nat. Chem. 9, 420–425 (2017).
De Poli, M. et al. Conformational photoswitching of a synthetic peptide foldamer bound within a phospholipid bilayer. Science 352, 575–580 (2016).
Formaggio, F. et al. The first water-soluble 310-helical peptides. Chemistry 6, 4498–4504 (2000).
Zieleniewski, F., Woolfson, D. N. & Clayden, J. Automated solid-phase concatenation of Aib residues to form long, water-soluble, helical peptides. Chem. Commun. 56, 12049–12052 (2020).
Woolfson, D. N. Coiled-coil design: updated and upgraded. Subcell. Biochem. 82, 35–61 (2017).
Fletcher, J. M. et al. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 1, 240–250 (2012).
Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 (1993).
Kumar, P. & Woolfson, D. N. Socket2: a program for locating, visualising, and analysing coiled-coil interfaces in protein structures. Bioinformatics 37, 4575–4577 (2021).
Swanson, C. J. & Sivaramakrishnan, S. Harnessing the unique structural properties of isolated alpha-helices. J. Biol. Chem. 289, 25460–25467 (2014).
Brown, R. A., Marcelli, T., De Poli, M., Sola, J. & Clayden, J. Induction of unexpected left-handed helicity by an N-terminal L-amino acid in an otherwise achiral peptide chain. Angew. Chem. Int. Ed. 51, 1395–1399 (2012).
Thomson, A. R. et al. Computational design of water-soluble alpha-helical barrels. Science 346, 485–488 (2014).
Thomas, F. et al. De novo-designed alpha-helical barrels as receptors for small molecules. ACS Synth. Biol. 7, 1808–1816 (2018).
Enkhbayar, P., Hikichi, K., Osaki, M., Kretsinger, R. H. & Matsushima, N. 310-helices in proteins are parahelices. Proteins 64, 691–699 (2006).
Kumar, P. & Bansal, M. HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J. Biomol. Struct. Dyn. 30, 773–783 (2012).
Lupas, A. N. & Gruber, M. The structure of alpha-helical coiled coils. Adv. Protein Chem. 70, 37–78 (2005).
Hunter, C. A. & Sanders, J. K. M. The nature of π-π interactions. J. Am. Chem. Soc. 112, 5525–5534 (1990).
Mortenson, D. E. et al. High-resolution structures of a heterochiral coiled coil. Proc. Natl Acad. Sci. USA 112, 13144–13149 (2015).
Fox, R. O. Jr. & Richards, F. M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature 300, 325–330 (1982).
Bunkoczi, G., Schiell, M., Vertesy, L. & Sheldrick, G. M. Crystal structures of cephaibols. J. Pept. Sci. 9, 745–752 (2003).
Mendel, D., Ellman, J. & Schultz, P. G. Protein biosynthesis with conformationally restricted amino acids. J. Am. Chem. Soc. 115, 4359–4360 (1993).
Leonard, D. J., Ward, J. W. & Clayden, J. Asymmetric α-arylation of amino acids. Nature 562, 105–109 (2018).
Collie, G. W. et al. Shaping quaternary assemblies of water-soluble non-peptide helical foldamers by sequence manipulation. Nat. Chem. 7, 871–878 (2015).
Wang, P. S. & Schepartz, A. β-Peptide bundles: Design. Build. Analyze. Biosynthesize. Chem. Commun. 52, 7420–7432 (2016).
Chandramouli, N. et al. Iterative design of a helically folded aromatic oligoamide sequence for the selective encapsulation of fructose. Nat. Chem. 7, 334–341 (2015).
Girvin, Z. C., Andrews, M. K., Liu, X. & Gellman, S. H. Foldamer-templated catalysis of macrocycle formation. Science 366, 1528–1531 (2019).
Wang, G. & Dunbrack, R. L. Jr. PISCES: a protein sequence culling server. Bioinformatics 19, 1589–1591 (2003).
Joosten, R. P. et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 39, D411–419 (2011).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
The PyMOL Molecular Graphics System Open-Source v2.4.0 (Schrödinger, 2021).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Laue, T., Shah, B., Ridgeway, T. & Pelletier, S. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Harding, S. E. et al.) 90–125 (Royal Society of Chemistry, 1992).
Zhao, H., Piszczek, G. & Schuck, P. SEDPHAT–a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Sheldrick, G. M. SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. A 71, 3–8 (2015).
Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42, 339–341 (2009).
Sammito, M. et al. ARCIMBOLDO_LITE: single-workstation implementation and use. Acta Crystallogr. D 71, 1921–1930 (2015).
Caballero, I. et al. ARCIMBOLDO on coiled coils. Acta Crystallogr. D 74, 194–204 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014).
Acknowledgements
P.K. and D.N.W. are supported by Biotechnology and Biological Sciences Research Council (BB/R00661X/1) and European Research Council (340764) grants to D.N.W. D.N.W. is also supported by BrisSynBio, a Biotechnology and Biological Sciences Research Council/Engineering and Physical Sciences Research Council (EPSRC)-financed Synthetic Biology Research Centre (BB/L01386X/1), and a Royal Society Wolfson Research Merit Award (WM140008). J.C. is supported by the European Research Council Advanced Grant DOGMATRON (agreement no. 884786) and an EPSRC Programme Grant (EP/P027067/1). We thank the University of Bristol School of Chemistry Mass Spectrometry Facility for access to the EPSRC-financed Bruker Ultraflex MALDI-TOF/TOF instrument (EP/K03927X/1), and BrisSynBio for access to peptide synthesizers. We thank C. Williams for collecting one-dimensional 1H nuclear magnetic resonance spectra. We thank Diamond Light Source for access to beamlines I03, I04, I04-1 and I24 (Proposal 23269) and M. Warren from I19 who helped N.G.P. with the direct methods solution. We thank T. Yeates (University of California, Los Angeles), K. Gupta, C. Tölzer, F. Zieleniewski and members of the Clayden and Woolfson laboratories and BrisSynBio for helpful discussions.
Author information
Authors and Affiliations
Contributions
P.K., J.C. and D.N.W. conceived the project. P.K. and D.N.W. designed the bioinformatics analyses, which were performed by P.K. P.K. and D.N.W. designed the sequences, which were synthesized, characterized and crystallized by P.K. P.K. and N.G.P. solved the X-ray crystal structures. P.K., J.C. and D.N.W. wrote the manuscript, which was read by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Giovanna Ghirlanda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
This file contains the following seven sections: Section 1, Bioinformatics analyses; Section 2, Analytical high-pressure liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization - time of flight (MALDI-TOF); Section 3, Circular dichroism (CD) spectroscopy; Section 4, Analytical ultracentrifugation (AUC); Section 5, DPH-binding analyses; Section 6, Structural analyses of 310-helix bundle; Section 7, Tables.
Supplementary Code
This file contains customized scripts used for bioinformatic analyses.
Rights and permissions
About this article
Cite this article
Kumar, P., Paterson, N.G., Clayden, J. et al. De novo design of discrete, stable 310-helix peptide assemblies. Nature 607, 387–392 (2022). https://doi.org/10.1038/s41586-022-04868-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04868-x
This article is cited by
-
Towards glycan foldamers and programmable assemblies
Nature Reviews Materials (2024)
-
Assembly of peptide nanostructures with controllable sizes
Nano Research (2024)
-
Stapling strategy for slowing helicity interconversion of α-helical peptides and isolating chiral auxiliary-free one-handed forms
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.