Abstract
µ-Opioid peptide receptor (MOPR) stimulation alters respiration, analgesia and reward behaviour, and can induce substance abuse and overdose1,2,3. Despite its evident importance, the endogenous mechanisms for MOPR regulation of consummatory behaviour have remained unknown4. Here we report that endogenous MOPR regulation of reward consumption in mice acts through a specific dorsal raphe to nucleus accumbens projection. MOPR-mediated inhibition of raphe terminals is necessary and sufficient to determine consummatory response, while select enkephalin-containing nucleus accumbens ensembles are engaged prior to reward consumption, suggesting that local enkephalin release is the source of the endogenous MOPR ligand. Selective modulation of nucleus accumbens enkephalin neurons and CRISPR–Cas9-mediated disruption of enkephalin substantiate this finding. These results isolate a fundamental endogenous opioid circuit for state-dependent consumptive behaviour and suggest alternative mechanisms for opiate modulation of reward.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The behavioural dataset supporting the current study is available from the author upon request. Source data are provided with this paper.
Code availability
Custom MATLAB analysis and code was created to appropriately organize, process, and combine photometry and single-photon recording data with associated behavioural data. Analysis code for photometry and single-photon imaging from Figs. 2, 4 will be made available on Github (https://github.com/BruchasLab).
References
Centers for Disease Control and Prevention. CDC grand rounds: prescription drug overdoses—a U.S. epidemic. Morb. Mortal. Wkly. Rep. 61, 10–13 (2012).
Volkow, N., Benveniste, H. & McLellan, A. T. Use and misuse of opioids in chronic pain. Annu. Rev. Med. 69, 451–465 (2018).
Wei, A. D. & Ramirez, J.-M. Presynaptic mechanisms and KCNQ potassium channels modulate opioid depression of respiratory drive. Front. Physiol. 10, 1407 (2019).
Kelley, A. E., Baldo, B. A. & Pratt, W. E. A proposed hypothalamic-thalamic-striatal axis for the integration of energy balance, arousal, and food reward. J. Comp. Neurol. 493, 72–85 (2005).
Mattison, J. The treatment of the morphine-disease. Indian Med. Gaz. 26, 65–68 (1891).
Herkenham, M. & Pert, C. B. In vitro autoradiography of opiate receptors in rat brain suggests loci of ‘opiatergic’ pathways. Proc. Natl Acad. Sci. USA 77, 5532–5536 (1980).
Massaly, N., Morón, J. A. & Al-Hasani, R. A trigger for opioid misuse: chronic pain and stress dysregulate the mesolimbic pathway and kappa opioid system. Front. Neurosci. 10, 480 (2016).
Castro, D. C. & Bruchas, M. R. A motivational and neuropeptidergic hub: anatomical and functional diversity within the nucleus accumbens shell. Neuron 102, 529–552 (2019).
Hughes, J. et al. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature 258, 577–580 (1975).
Simantov, R. & Snyder, H. Isolation and structure identification of a morphine-like peptide ‘enkephalin’ in bovine brain. Life Sci. 18, 781–787 (1976).
Bakshi, V. P. & Kelley, A. E. Feeding induced by opioid stimulation of the ventral striatum: role of opiate receptor subtypes. J. Pharmacol. Exp. Ther. 265, 1253–1260 (1993).
Resendez, S. L. et al. μ-Opioid receptors within subregions of the striatum mediate pair bond formation through parallel yet distinct reward mechanisms. J. Neurosci. 33, 9140–9149 (2013).
Castro, D. C. & Berridge, K. C. Opioid hedonic hotspot in nucleus accumbens shell: mu, delta, and kappa maps for enhancement of sweetness ‘liking’ and ‘wanting’. J. Neurosci. 34, 4239–4250 (2014).
Cui, Y. et al. Targeted expression of μ-opioid receptors in a subset of striatal direct-pathway neurons restores opiate reward. Nat. Neurosci. 17, 254–261 (2014).
Bodnar, R. J., Glass, M. J., Ragnauth, A. & Cooper, M. L. General, µ and κ opioid antagonists in the nucleus accumbens alter food intake under deprivation, glucoprivic and palatable conditions. Brain Res. 700, 205–212 (1995).
Kelley, A. E., Bless, E. P. & Swanson, C. J. Investigation of the effects of opiate antagonists infused into the nucleus accumbens on feeding and sucrose drinking in rats. J. Pharmacol. Exp. Ther. 278, 1499–1507 (1996).
Kramer, T. H. et al. Novel peptidic mu opioid antagonists: pharmacologic characterization in vitro and in vivo. J. Pharmacol. Exp. Ther. 249, 544–551 (1989).
Tervo, D. G. R. et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016).
Zhao, Z.-Q. et al. Central serotonergic neurons are differentially required for opioid analgesia but not for morphine tolerance or morphine reward. Proc. Natl Acad. Sci. USA 104, 14519–14524 (2007).
Land, B. B. et al. Activation of the kappa opioid receptor in the dorsal raphe nucleus mediates the aversive effects of stress and reinstates drug seeking. Proc. Natl Acad. Sci. USA 106, 19168–19173 (2009).
Nectow, A. R. et al. Identification of a brainstem circuit controlling feeding. Cell 170, 429-442.e11 (2017).
Huang, K. W. et al. Molecular and anatomical organization of the dorsal raphe nucleus. eLife 8, e46464 (2019).
Gunaydin, L. A. et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014).
Siuda, E. R. et al. Spatiotemporal control of opioid signaling and behavior. Neuron 86, 923–935 (2015).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Hunker, A. C. et al. Conditional single vector CRISPR/SaCas9 viruses for efficient mutagenesis in the adult mouse nervous system. Cell Rep. 30, 4303-4316.e6 (2020).
Al-Hasani, R. et al. In vivo detection of optically-evoked opioid peptide release. eLife 7, e36520 (2018).
Maldonado-Irizarry, C. S., Swanson, C. J. & Kelley, A. E. Glutamate receptors in the nucleus accumbens shell control feeding behavior via the lateral hypothalamus. J. Neurosci. 15, 6779–6788 (1995).
Vachez, Y. M. et al. Ventral arkypallidal neurons inhibit accumbal firing to promote reward consumption. Nat. Neurosci. 24, 379–390 (2021).
Castro, D. C., Terry, R. A. & Berridge, K. C. Orexin in rostral hotspot of nucleus accumbens enhances sucrose ‘liking’ and intake but scopolamine in caudal shell shifts ‘liking’ toward ‘disgust’ and ‘fear’. Neuropsychopharmacology 41, 2101–2111 (2016).
Georgescu, D. et al. The hypothalamic neuropeptide melanin-concentrating hormone acts in the nucleus accumbens to modulate feeding behavior and forced-swim performance. J. Neurosci. 25, 2933–2940 (2005).
Lim, B. K., Huang, K. W., Grueter, B. A., Rothwell, P. E. & Malenka, R. C. Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature 487, 183–189 (2012).
O’Connor, E. C. et al. Accumbal D1R neurons projecting to lateral hypothalamus authorize feeding. Neuron 88, 553–564 (2015).
Berthoud, H.-R. & Münzberg, H. The lateral hypothalamus as integrator of metabolic and environmental needs: from electrical self-stimulation to opto-genetics. Physiol. Behav. 104, 29–39 (2011).
Corre, J. et al. Dopamine neurons projecting to medial shell of the nucleus accumbens drive heroin reinforcement. eLife 7, e39945 (2018).
Di Chiara, G. & Imperato, A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc. Natl Acad. Sci. USA 85, 5274–5278 (1988).
Smith, K. S. & Berridge, K. C. The ventral pallidum and hedonic reward: neurochemical maps of sucrose ‘liking’ and food intake. J. Neurosci. 25, 8637–8649 (2005).
Land, B. B. et al. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J. Neurosci. 28, 407–414 (2008).
Zelikowsky, M. et al. The neuropeptide Tac2 controls a distributed brain state induced by chronic social isolation stress. Cell 173, 1265-1279.e19 (2018).
Parker, K. E. et al. A paranigral VTA nociceptin circuit that constrains motivation for reward. Cell 178, 653-671.e19 (2019).
Al-Hasani, R. et al. Distinct subpopulations of nucleus accumbens dynorphin neurons drive aversion and reward. Neuron 87, 1063–1077 (2015).
Massaly, N. et al. Pain-induced negative affect is mediated via recruitment of the nucleus accumbens kappa opioid system. Neuron 102, 564–573.e6 (2019).
Fetterly, T. L. et al. Insulin bidirectionally alters NAc glutamatergic transmission: interactions between insulin receptor activation, endogenous opioids, and glutamate release. J. Neurosci. 41, 2360–2372 (2021).
Schmid, C. L. et al. Bias factor and therapeutic window correlate to predict safer opioid analgesics. Cell 171, 1165-1175.e13 (2017).
Singh, J. & Desiraju, T. Differential effects of opioid peptides administered intracerebrally in loci of self-stimulation reward of lateral hypothalamus and ventral tegmental area–substantia nigra. NIDA Res. Monogr. 87, 180–191 (1988).
Lemos, J. C., Roth, C. A. & Chavkin, C. Signaling events initiated by kappa opioid receptor activation: quantification and immunocolocalization using phospho-selective KOR, p38 MAPK, and KIR 3.1 antibodies. Methods Mol. Biol. 717, 197–219 (2011).
Banghart, M. R. & Sabatini, B. L. Photoactivatable neuropeptides for spatiotemporally precise delivery of opioids in neural tissue. Neuron 73, 249–259 (2012).
Banala, S. et al. Photoactivatable drugs for nicotinic optopharmacology. Nat. Methods 15, 347–350 (2018).
Zhang, Y. et al. Battery-free, lightweight, injectable microsystem for in vivo wireless pharmacology and optogenetics. Proc. Natl Acad. Sci. USA 116, 21427–21437 (2019).
Patriarchi, T. et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018).
Sun, F. et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481-496.e19 (2018).
Wang, F. et al. RNAscope. J. Mol. Diagn. 14, 22–29 (2012).
Matthes, H. W. et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the µ-opioid-receptor gene. Nature 383, 819–823 (1996).
McCall, J. G. et al. CRH engagement of the locus coeruleus noradrenergic system mediates stress-induced anxiety. Neuron 87, 605–620 (2015).
Resendez, S. L. et al. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat. Protoc. 11, 566–597 (2016).
Zhou, P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018).
Acknowledgements
We thank L. Lawson, D. Blumenthal, M. Chung, T. Hobbs and C. Pizzano for animal colony maintenance; the Bruchas laboratory, Stuber laboratory and multiple trainees and faculty within the NAPE Center for helpful discussions; B. Kieffer for the Oprm1fl/fl mice; and G. Scherrer for the Penk-Cre mice. D.C.C. was funded by NIH grants NS007205, DA043999, DA049862 and DA051489. C.E.P. was funded by NIH grant DA051124. M.A.R. was funded by NIH grant DK121883 and a NARSAD Young Investigator Award. J.A.M. was funded by NIH grants DA041781, DA042499 and DA045463. G.D.S. was funded by DA032750, DA038168 and DA048736. M.R.B. was funded by NIH grants R37DA033396 and R61DA051489 and the Mallinckrodt Endowed Professorship. M.R.B., G.D.S. and L.S.Z. were funded by NIH grant P30DA048736.
Author information
Authors and Affiliations
Contributions
D.C.C. designed and performed experiments, collected and analysed data, and wrote the manuscript. C.S.O., E.T.Z. and A.G. performed experiments and collected data. M.A.R. collected and analysed electrophysiological data. A.C.H. and L.S.Z. designed and analysed CRISPR virus. C.E.P designed and analysed fibre photometry data. S.C.P. designed and analysed one-photon data. M.R.B. and J.A.M. facilitated resources for generation of Oprm1fl/fl × Penk and Oprm1fl/fl × Pdyn mouse lines. J.A.M., L.S.Z. and G.D.S. helped to design experiments, discuss results and write the manuscript. M.R.B. helped lead the design, analysis, oversight of experiments, discuss results, provide resources and write the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Brigitte Kieffer, Christian Lüscher and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Endogenous MOPR activation in mNAcSh is necessary for potentiating state-dependent consummatory behavior.
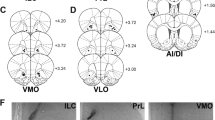
a. Placement map of each microinjector tip (blue = intake suppression compared to vehicle-deprived, green = intake enhancement compared to vehicle-deprived). b. Schematic of vehicle (ACSF, gray, top) or drug (CTAP, blue, bottom) microinjections into areas surrounding nucleus accumbens (NAc) medial shell. c. CTAP (blue) had no effect on ad libitum or hunger enhanced intake compared to vehicle (gray) control days when injections were outside NAc medial shell (n = 8). d. In situ hybridization of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200µm). e, f. Quantification of MOPR expression in mNAcSh. g. Schematic of Oprm1fl/fl x Pdyn-Cre mouse line cross. h, i. In situ hybridization (h) and quantification (i) of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200 µm) in Oprm1fl/fl x Pdyn-Cre mouse line. j. Loss of MOPRs on Pdyn-Cre+ neurons did not disrupt normal ad libitum or food deprived enhanced intake compared to Pdyn-Cre- littermate control mice (n = 9 Cre-, 10 Cre+). k and l. In situ hybridization (k) and quantification (l) of Pdyn, Penk and Oprm1 in NAc medial shell (scale bar = 200 µm) in Oprm1fl/fl x Penk-Cre mouse line. m. Schematic and image of rAAV5-CMV-Cre-GFP injections into NAc medial shell of Oprm1fl/fl mice. n. Schematic of combined viral spread map of local MOPR deletion. o. Schematic (top) and image (bottom) of AAV2retro-CMV-myc-NLS-Cre or AAV2retro-GFP-Cre injections into NAc (left); retrogradely labeled cells in dorsal raphe nucleus (right). All error bars represent ± SEM and n = biologically independent mice or cells (f, i, l). Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, j).
Extended Data Fig. 2 Endogenous MOPR activation in mNAcSh is necessary for potentiating state-dependent avoidance behavior.
a. Schematic of elevated zero maze (EZM) test. Mice were tested after habituation to the test room (Unrestrained) or after 30 min of restraint stress (Restraint). b. Example heat plots of time spend in the open arms of the EZM in wildtype (WT, left) or Oprm1 KO (KO, right) after no restraint (top) or 30 min of restraint (bottom). c. Unrestrained WT mice spent ~30% of the EZM test in the open arms. Mice exposed to restraint stress significantly reduced their exploration to 10%. Pretreatment with naloxone prevented Restraint induced avoidance. Oprm1 KO mice did not display open arm avoidance after Restraint. Penk-Cre x cKO mice displayed normal avoidance of the open arms after Restraint (n = 9 WT Unrestrained, 9 WT Restrained, 8 Oprm1 KO Unrestrained, 7 Oprm1 KO Restrained, 8 Oprm1fl/fl x Penk-Cre- Restrained, 9 Oprm1fl/fl x Penk-Cre+ Restrained). d. Schematic of food intake assay after food deprivation or Restraint. e. Food deprived mice showed normal increase in intake relative to their ad libitum test day. Mice exposed to Restraint did not increase food intake relative to their Unrestrained test day (n = 11 deprived, 10 Restrained). All error bars represent ± SEM and n = biologically independent mice. Medians marked with orange bar. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, e).
Extended Data Fig. 3 LDRNPenk-NAc projections express MOPRs.
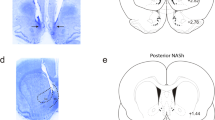
a. Retrograde fluorescently tagged cells in amygdala of Penk-Cre+ mouse after injections into nucleus accumbens medial shell. b. (left) In situ hybridization of Oprm1, Slc32a1 and Slc17a6 in DRN (scale bar = 200 µm). Zoomed in and channel separated images (right) of the red square in the left panel. c. Quantification of in situ from panel b. d, e. Schematic and image of a local injection of AAV2retro-GFP-Cre into nucleus accumbens shell (scale bar = 200 µm). Zoomed in images of e are designated as red (i) and yellow (ii) boxes. f. (left) In situ hybridization of Oprm1, Penk and Cre in dorsal raphe nucleus (scale bar = 200 µm). Zoomed in and channel separated images (right) of the red square in the left panel. g. Quantification of in situ from panel f. h. Schematic of CTb experiment. i. Fluorescently tagged CTb was injected into mNAcSh (scale bar = 200 µm). j. CTb tagged cells were observed in dorsal raphe nucleus, including the lateral sites in which enkephalin neurons were labeled in Fig. 2a (scale bar = 200 µm). Red square shows zoomed in image (right) with labeled cells (depicted by white arrows). All error bars represent ± SEM and n = biologically independent cells.
Extended Data Fig. 4 Non-DRN sites do not mediate food deprived potentiation of sucrose consumption.
a. Local MOPR deletion in paraventricular thalamus of Oprm1fl/fl mice did not reduce food deprived enhanced intake (n = 8, paired t-test t(7) = 9.634, p < 0.001). b. Schematic of local caspase ablations in either ventral pallidum or basomedial amygdala. c. Local and cell-type specific ablation of enkephalin neurons in VP or BMA did not reduce food deprived enhanced intake relative to Cre- control mice (n = 8 Cre-, 9 VP, 7 BMA). d. Caspase injection site confirmation in ventral pallidum (scale bar = 200 µm). AAV2-FLEX-taCas3-TEPp and AAV5-hsyn-EYFP were coinfused for cell-type specific deletion, and non-specific labeling.e, f. Schematic and image of a local injection of AAV5-Ef1a-DIO-EYFP into DRNPenk (scale bar = 200 µm). g. Image of dorsal raphe projection fibers from e (scale bar = 200 µm). h. oEPSC amplitude was reduced by the application of DNQX (n = 8). Blue shaded region indicates duration of optical stimulation. i. oIPSC amplitude was reduced by the application of gabazine (n = 5). All error bars represent ± SEM and n = biologically independent mice or cells (h, i). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c) or two tailed paired t-test (h, i).
Extended Data Fig. 5 LDRNMOPR-mNAcSh projection activity is negatively modulated by sucrose consumption in an opioid receptor dependent manner.
a. Schematic of photometry and voluntary sucrose consumption paradigm. b. Expression of GCaMP6s in DRNPenk (left, scale bar = 200 µm) and fiber placement in mNAcSh (scale bar = 200 µm). c. FD increases food intake (green) relative to ad libitum intake (white), and is reduced by systemic naloxone (blue, n = 8). d. Eating microstructure across ad libitum/saline (white), food deprived/saline (green) and food deprived/naloxone (blue) test days. After food deprivation, the majority of sucrose pellets consumed during multi-pellet bouts (left), and more pellets are eaten per multi-pellet bout (right). This shift in eating behavior is blunted by naloxone. e. Example of raw 405nm and 470nm channels from photometry experiments. f. Example of raw df/f trace highlighting specific single pellet (black) or multi-pellet (green) intake events during food deprived test day. Colored boxes on top expand color matched portions of the trace below. Orange lines indicate onset of pellet consumption. g. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh terminals in FD/saline condition (green) and FD/naloxone condition (blue). h. Average Z-scored trace (dark line) and error in on the ad libitum test day. When GCaMP activity is aligned to the onset of each pellet eaten in the ad libitum condition, no significant deviation in activity is observed (black, top). When aligned to only multi-pellet bout onset, there is no deviation from baseline activity (brown, bottom). i. Schematic of regulated food intake paradigm. Two pellets were non-contingently delivered every 90-150 s for 30 min. Mice were tested in either FD/saline or FD/naloxone conditions. j. Total number of sucrose pellets eaten in the regulated intake paradigm. Mice ate significantly more pellets on the food deprivation/saline test day (green), which was reduced to baseline levels after systemic naloxone (blue) (n = 10) k. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh. Food deprived/saline trace (green) shows rapid and sustained inhibition. Food deprived/naloxone trace (blue) shows blunted response. l. Heatmap of individual trials across all tested mice in food deprived/saline (green) or food deprived/naloxone test days. Orange lines indicate onset of pellet consumption. m. Quantification of the average Z-score twenty seconds prior to the onset of multi-pellet pellet bouts versus twenty seconds after the onset. FD/saline traces (green) show significant reductions in GCaMP6s activity whereas FD/naloxone traces (blue) do not (n = 10). All error bars represent ± SEM and n = biologically independent mice. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, j, m).
Extended Data Fig. 6 MOPR activation on LDRNMOPR-mNAcSh is sufficient to enhance consummatory behavior.
a. Schematic of the Tail Immersion Test. Mice were tested at time 0, were injected with morphine (5mg/kg, s.c.), then tested every 10 min for up to 60 min, then again at 90 min. b. WT, Oprm1 KO, and Oprm1 KO x Penk-Cre rescue mice all show similar baseline responses at time 0. After morphine administration, WT mice significantly increase their latency to flick their tail, whereas Oprm1 KO and Oprm1 KO x Penk-Cre show no analgesic response to morphine mice (n = 5 WT, 2 Oprm1 KO, 6 Oprm1 KO x Penk-Cre rescue; WT/T0 vs Oprm1 KO/T0 p = 0.116, WT/T0 vs Oprm1 KO x Penk-Cre rescue/T0 p = 0.063, WT/T10 vs Oprm1 KO/T10 p < 0.001, WT/T10 vs Oprm1 KO x Penk-Cre rescue/T10 p < 0.001, WT/T90 vs Oprm1 KO/T90 p < 0.001, WT/T90 vs Oprm1 KO x Penk-Cre rescue/T90 p < 0.001). Statistical differences between WT and Oprm1 KO designated as (***) and differences between WT and Oprm1 KO x Penk-Cre rescue designated as (+++). c. Raster plots of individual licking events for one mouse, separated by trial. Only trials in which mice licked were included. d. Schematic of ChR2 experiments. e. Expression of EYFP-tagged ChR2 in LDRNPenk cell bodies (left, scale bar = 200 µm) and fiber placement in mNAcSh (right, scale bar = 200 µm). f. Penk-Cre- and Penk-Cre+ mice licked similar amounts for a sucrose solution in the ad libitum/No Laser condition. ChR2 photo-stimulation did not reduce licking (n = 7 Cre-, 6 Cre+). g. Penk-Cre- and Penk-Cre+ mice licked similar amounts for a sucrose solution in the WD/No Laser condition. By contrast, ChR2 photo-stimulation significantly reduced Cre+ licking, but not Cre- licking. h. Raster plots of individual licking events for one Penk-Cre+ mouse in the WD condition, separated by trial. Only trials in which mice licked were included. ChR2 photo-stimulation disrupted lick bout behavior compared to No Laser test days. All error bars represent ± SEM and n = biologically independent mice or cells (a). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, f, g).
Extended Data Fig. 7 mNAcSh enkephalinergic ensembles are modulated by physiological state and potentiate consummatory behavior.
a. Sucrose consumed during ad libitum and food deprived test days (n = 5). Miniscope headmount did not disrupt normal intake behaviors. b. Examples of individual cell traces aligned to initiation of a multi-pellet bout. c. Average trace of all tracked cells aligned to bout consumption on food deprived test day. d. TSNE plot of clusters for multi-pellet bouts during the ad libitum state. e. Average trace of Onset activated neurons (cluster 1). f. Average trace of Pre-onset activated neurons (cluster 2). g. Total proportion and overlap of enkephalin neuron subpopulations modulated by multi-pellet bouts (pink), food sniffs (orange), rearing (blue), and grooming (brown). h. TSNE (left) and mean Z-scored traces (right) of food sniffing behavior sorted by kmeans clustering. i. TSNE (left) and mean Z-scored traces (right) of rearing behavior sorted by kmeans clustering. j. TSNE (left) and mean Z-scored traces (right) of grooming behavior sorted by kmeans clustering. All error bars represent ± SEM (a) or SEM is represented by the shaded region surrounding the trace (c, e, f, h, i, j).
Extended Data Fig. 8 Modulation of mNAcSh or POMC-containing neurons during sucrose consumption.
a. Schematic of hM3D(Gi) (left) and hM3D(Gq) (right) DREADD experiments. b. Fluorescent micrograph of mCherry-tagged enkephalin cells in mNAcSh for hM3D(Gi) (left) and hM3D(Gq) (right) experiments (scale bar = 200µm). c. CNO injections suppressed hunger enhanced intake in Cre+ mice, but had no effect in Cre- mice (n = 15 Cre-, 12 Cre+). d. CNO injections increased intake above the already elevated food deprived intake in Penk-Cre+ mice, but had no effect in Penk-Cre- mice (n = 7 Cre-, 10 Cre+). e. Systemic CNO administration (3mg/kg, i.p.) suppressed the already low ad libitum intake in Penk-Cre+ mice, but did not reduce intake in Penk-Cre- mice. f. Systemic CNO administration (3mg/kg, i.p.) had no effect on ad libitum intake in Penk-Cre- or Penk-Cre+ mice). g. Gq or Gi DREADD injections into arcuate nucleus of POMC-Cre mice. h. Micropictograph of mCherry-tagged, DREADD-expressing cells in arcuate nucleus (scale bar = 200 μm). i–k. Neither Gi nor Gq stimulation had an effect on ad libitum or food deprived intake in Cre- or Cre+ mice (n = 7 Cre-, 7 Gq, 9 Gi). All error bars represent ± SEM and n = biologically independent mice. Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (c, d, e, f, i, j, k).
Extended Data Fig. 9 Endogenous mu-opioid peptide within mNAcSh is necessary for potentiating consummatory behavior.
a. FISH (scale bar = 200 µm) of mNAcSh caspase injections. b. FISH quantification of (a). c. FISH (scale bar = 200 µm) of mNAcSh CRISPR injections. d. FISH quantification of (c). e. Schematic of CRISPR virus development and validation. f. Sequencing of GFP+ nuclei: (Top) sgPenk sequence with PAM underlined and SaCas9 cut site indicated by black arrow. (Middle) Sanger sequencing results displaying multiple peaks beginning at the SaCas9 predicted cut site. (Bottom) Top ten mutations at cut site with the percent of occurrence on the left. Insertions: underlined. Deletions: marked with “-“. Affected sites after SaCas9 insertion: shaded brown. g. Percent of wild type (black), deletions (brown), insertions (pink), and base changes (white) as percent of total reads for GFP+ and GFP- nuclei. h. Frequency distribution of insertions (pink) and deletions (brown) for Penk from GFP+ nuclei. i. Sanger sequencing results displaying no unusual peaks beginning at the SaCas9 predicted cut site. j. Unilateral hits or bilateral misses of NAc medial shell with the CRISPR mediated deletion of enkephalin did not reduce food deprived enhanced intake (n = 4). k. Fluorescent micrograph of CRISPR virus expression in dorsal raphe nucleus. l. Deletion of enkephalin from dorsal raphe nucleus did not reduce food deprived enhanced intake (n = 8) m. Deletion of dynorphin in NAc medial shell did not reduce food deprived enhanced intake (n = 7). n. Heatmap of individual trials across all tested mice in CRISPR/saline (orange) or Control/saline (gray) test days. Orange lines indicate onset of pellet consumption. o. Average Z-scored trace aligned to onset of multi-pellet consumption of LDRNPenk-mNAcSh after systemic naloxone injections. Control-treated mice (blue) show blunted inhibition. CRISPR-expressing trace (brown) shows negligible and phasic inhibition. p. Heatmap of individual trials across all tested mice in CRISPR/naloxone (orange) or Control/naloxone (blue) test days. q. Quantification of the average Z-score twenty seconds prior to the onset of multi-pellet pellet bouts versus twenty seconds after the onset. Neither Control traces (blue) nor CRISPR traces (brown) show significant deviations in GCaMP activity. Some mice did not lick on naloxone treated days and were therefore not included in this analysis (CRIPSR n = 3, Control n = 3). All error bars represent ± SEM and n = biologically independent mice or cells (b, d). Post hoc p-values are derived from Two-way ANOVA with Sidak multiple comparisons (q) or two-tailed paired t-test (j, l, m).
Extended Data Fig. 10 A LDRNMOPR-mNAcShPenk circuit mediates potentiation of consummatory behaviors.
Schematic of voluntary sucrose consumption task (upper left) and LDRN-mNAcSh projection (upper right). Effects of LDRNMOPR-mNAcShPenk manipulations on behavior (lower left) and schematic of hypothesized physiology (lower right).
Supplementary information
Supplementary Discussion
This text considers how the data described in this manuscript are contextualized by known and unknown information. It examines potential circuits, psychological functions, and intracellular signalling mechanisms of interest.
Supplementary Video 1
This video demonstrates endoscopic recording of GCaMP6s labelled Penk-cre neurons in mNAcSh in an awake and behaving mouse. Behavioural and imaging videos are temporally synchronized.
Rights and permissions
About this article
Cite this article
Castro, D.C., Oswell, C.S., Zhang, E.T. et al. An endogenous opioid circuit determines state-dependent reward consumption. Nature 598, 646–651 (2021). https://doi.org/10.1038/s41586-021-04013-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04013-0
This article is cited by
-
In vivo photopharmacology with a caged mu opioid receptor agonist drives rapid changes in behavior
Nature Methods (2023)
-
Lateral hypothalamic proenkephalin neurons drive threat-induced overeating associated with a negative emotional state
Nature Communications (2023)
-
Human OPRM1 and murine Oprm1 promoter driven viral constructs for genetic access to μ-opioidergic cell types
Nature Communications (2023)
-
Local modulation by presynaptic receptors controls neuronal communication and behaviour
Nature Reviews Neuroscience (2022)
-
An opioid-gated thalamoaccumbal circuit for the suppression of reward seeking in mice
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.