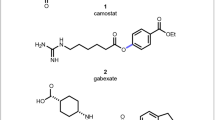
Abstract
Chemical transformations determine the structure of a product, and therefore its properties, which in turn affect complex macroscopic functions such as the metabolic stability of pharmaceuticals or the volatility of perfumes. Therefore, reaction selection can influence the success or failure of a candidate molecule to meet a functional objective. The coupling of an amine with a carboxylic acid to form an amide bond is the most popular chemical reaction used for drug discovery1. However, there are many other ways to connect these two common functional groups together. Here we show computationally that amines and acids can couple via hundreds of hypothetical yet plausible transformations, and we demonstrate experimentally the application of a dozen such reactions. To investigate the contribution of chemical transformations to properties, we developed a string-based notation and used an enumerative combinatorics approach to produce a map of conceivable amine–acid coupling transformations, which can be charted using chemoinformatic techniques. We find that critical physicochemical parameters of the products, such as partition coefficient and polar surface area, vary considerably depending on the transformation chosen. Data mining the amine–acid coupling system produced here should enable reaction discovery, which we demonstrate by developing an esterification reaction found within the mapped space. Complex molecules with distinct property profiles can also be discovered within the amine–acid coupling system, as we show here via the late-stage diversification of drugs and natural products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data, including experimental details, spectral data and raw .fid files generated or analysed during this study are included in Supplementary Information. Chemoinformatic data are available at https://github.com/cernaklab/acid-amine-enumeration.
Code availability
All code produced during this study can be found at https://github.com/cernaklab/acid-amine-enumeration.
References
Boström, J. et al. Expanding the medicinal chemistry synthetic toolbox. Nat. Rev. Drug Discov. 17, 709–727 (2018); correction 17, 922 (2018).
Liu, J. et al. Predicting organ toxicity using in vitro bioactivity data and chemical structure. Chem. Res. Toxicol. 30, 2046–2059 (2017).
Waring, M. J. et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14, 475–486 (2015).
Liu, R., Li, X. & Lam, K. S. Combinatorial chemistry in drug discovery. Curr. Opin. Chem. Biol. 38, 117–126 (2017).
Burke, M. D. & Schreiber, S. L. A planning strategy for diversity-oriented synthesis. Angew. Chem. Int. Ed. 43, 46–58 (2003).
Ugi, I. et al. New elements in the representation of the logical structure of chemistry by qualitative mathematical models and corresponding data structures. In Computer Chemistry (ed. Ugi, I.) 199–233 (Springer, 1993).
Gesmundo, N. J. et al. Nanoscale synthesis and affinity ranking. Nature 557, 228–232 (2018).
Buitrago Santanilla, A. et al. Nanomole-scale high-throughput chemistry for the synthesis of complex molecules. Science 347, 49–53 (2015).
Granda, J. M. et al. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature 559, 377–381 (2018); correction 570, E67–E69 (2019).
Perera, D. et al. A platform for automated nanomole-scale reaction screening and micromole-scale synthesis in flow. Science 359, 429–434 (2018).
Coley, C.W., et al., A robotic platform for flow synthesis of organic compounds informed by AI planning. Science 365, eaax1566 (2019).
Zahrt, A. F. et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science 363, eaau5631 (2019).
Ahneman, D. T. et al. Predicting reaction performance in C–N cross-coupling using machine learning. Science 360, 186–190 (2018).
Reid, J. P. & Sigman, M. S. Holistic prediction of enantioselectivity in asymmetric catalysis. Nature 571, 343–348 (2019).
McNally, A., Prier, C. K. & MacMillan, D. W. Discovery of an α-amino C–H arylation reaction using the strategy of accelerated serendipity. Science 334, 1114–1117 (2011).
Troshin, K. & Hartwig, J. F. Snap deconvolution: an informatics approach to high-throughput discovery of catalytic reactions. Science 357, 175–181 (2017).
Bickerton, G. R. et al. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Wager, T. T. et al. Moving beyond rules: the development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 1, 435–449 (2010).
Hill, A. P. & Young, R. J. Getting physical in drug discovery: a contemporary perspective on solubility and hydrophobicity. Drug Discov. Today 15, 648–655 (2010).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Fu, M. C. et al. Boron-catalyzed N-alkylation of amines using carboxylic acids. Angew. Chem. Int. Ed. 54, 9042–9046 (2015).
Alla, S. K., Sadhu, P. & Punniyamurthy, T. Organocatalytic syntheses of benzoxazoles and benzothiazoles using aryl iodide and oxone via C–H functionalization and C–O/S bond formation. J. Org. Chem. 79, 7502–7511 (2014).
Huang, L., Hackenberger, D. & Gooßen, L. J. Iridium-catalyzed ortho-arylation of benzoic acids with arenediazonium salts. Angew. Chem. Int. Ed. 54, 12607–12611 (2015).
Lipinski, C. A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44, 235–249 (2000).
Mao, R., Frey, A., Balon, J. & Hu, X. Decarboxylative C(sp 3)–N cross-coupling via synergetic photoredox and copper catalysis. Nat. Catal. 1, 120–126 (2018).
Acknowledgements
We thank the University of Michigan College of Pharmacy for start-up funds. S. McCarty is thanked for preliminary experiments and R. Zhang is thanked for discussions.
Author information
Authors and Affiliations
Contributions
B.M. performed chemoinformatic studies. Y.S., W.L., B.M. and T.C. performed chemistry experiments. All authors analysed the data. B.M. and T.C. wrote the manuscript. T.C. designed and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The University of Michigan has filed a patent on the technique described herein that lists T.C., B.M. and Y.S. as inventors.
Additional information
Peer review information Nature thanks Connor W. Coley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Workflow for enumeration of amine–acid transformations.
For a pair of coupling partners, we consider a reaction at the functional group A (amine) and B (carboxylic acid oxygen, B[O] or carbon, B[C]). Deamination reactions are noted as −A and decarboxylation reactions are noted as −B. Enumeration following steps 1–3 produces 320 transformations. For the enumeration of all syn- and anti-diastereomers (step 4), consult also Extended Data Fig. 6.
Extended Data Fig. 2 Kernel density estimate plots for 320 conceivable amine–acid coupling transformations.
Distribution of common physical properties from the achiral amine–acid coupling of ethylamine, ethenamine, propanoic acid and acrylic acid. MW, molecular weight; HBA, hydrogen bond acceptor; HBD, hydrogen bond donor; PSA, polar surface area; FSP3, fraction sp3; ROTB, rotatable bonds; FC, formal charge; QED, quantitative estimate of drug-likeness.
Extended Data Fig. 3 Number of DrugBank hits per transformation.
This bar chart shows how many times a transformation is found in the DrugBank database. Each number on the abscissa maps to a transformation listed in Extended Data Table 1.
Extended Data Fig. 4 Decarboxylative transformations from the enumeration scheme.
Decarboxylative reactions that produce an amine bound to an sp3 or sp2 carbon chain appear in high frequency. These reactions can be used to synthesize a large number of drugs contained in DrugBank. Each transformation can be found by its corresponding number in Extended Data Table 1. The colour scale is the same as in Fig. 2. rxn, reaction.
Extended Data Fig. 5 Transformations from the enumeration scheme found in specific drugs.
The chord diagrams show connectivity of transformation substructures as retrosynthetic disconnections in target molecules, with red and blue dots highlighting the transformations shown at left in each panel. a, Noscapine connects to 112 of the transformations. b, Quinine connects to 96 transformations. c, Sitagliptin connects to 55 transformations. The colour scale is the same as in Fig. 2.
Extended Data Fig. 6 Enumeration of regioisomers and diastereomers.
a, The transformation substructures enumerated in Fig. 3 are from the 320 achiral bond arrangements available from coupling 1, 2 and their sp2 variants ethenamine and acrylic acid. b, To sample three-dimensional and regiochemical space, a β′ substituent was added as a differentiating substituent. The β′ substituent may be any substituent, but is enumerated as being distinct from the β substituent. Considering this regiochemical enumeration increases the 320 achiral coupling transformations to 588. c, Subsequent enumeration of all possible diastereomers leads to 1,005 chiral coupling transformations. These 1,005 three-dimensional substructures were used as inputs in the principle moment of inertia plot in Extended Data Fig. 7.
Extended Data Fig. 7 Principal moment of inertia plot of 1,005 amine–acid coupling transformations incorporating stereochemistry and regiochemistry.
In this expanded three-dimensional space, regiochemistry and stereochemistry of the transformations were considered. A total of 1,005 ways to connect an amine to an acid were found. The products presented a diversity of properties and three-dimensional shapes. Each molecule is coloured by its quantitative estimate of drug-likeness.
Extended Data Fig. 8 High-throughput experimentation for the discovery of a copper-promoted esterification reaction.
a, An esterification reaction discovered through reaction screening of transition metals with ligands and additives. b, Recipe and well mapping. c, Calibration curve, for product 17 versus caffeine internal standard, used to convert the ultraperformance liquid chromatography with ultraviolet–visible spectrometry peak area to concentration, and thus to assay yield. Error bars show deviation among triplicate injections. d, Heat map depicting assay yield screening results. CuI with AgNO3 and pyridine showed the most promising results, achieving 18.5% assay yield using 30 mol% CuI with AgNO3.
Extended Data Fig. 9 Kernel density estimate plots of a series of complex molecules as substrates in the amine–acid coupling system.
The amine–acid pair depicted was used as an input to combinatorial enumeration, and the number of valid products identified is noted for each pairing. Distributions of common physical properties are shown for each coupling set. Abbreviations are as in Extended Data Fig. 2.
Supplementary information
Supplementary Information
This file contains a summary of methods, equipment, and techniques used to obtain the data presented in the manuscript. Furthermore, it contains an experiential section detailing instructions for all experiments performed and 42 NMR spectra for compounds presented in the main text. Finally, links are provided to GitHub repositories containing the fid files for all NMR spectra and the code used when generating chemoinformatic data and analyses.
Rights and permissions
About this article
Cite this article
Mahjour, B., Shen, Y., Liu, W. et al. A map of the amine–carboxylic acid coupling system. Nature 580, 71–75 (2020). https://doi.org/10.1038/s41586-020-2142-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2142-y
This article is cited by
-
Exploring the combinatorial explosion of amine–acid reaction space via graph editing
Communications Chemistry (2024)
-
Rapid planning and analysis of high-throughput experiment arrays for reaction discovery
Nature Communications (2023)
-
The isocyanide SN2 reaction
Nature Communications (2023)
-
Streamlining the synthesis of amides using Nickel-based nanocatalysts
Nature Communications (2023)
-
Miniaturization of popular reactions from the medicinal chemists’ toolbox for ultrahigh-throughput experimentation
Nature Synthesis (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.