Abstract
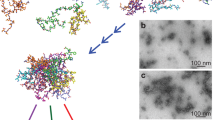
Protein crystallization is important in structural biology, disease research and pharmaceuticals. It has recently been recognized that nonclassical crystallization—involving initial formation of an amorphous precursor phase—occurs often in protein, organic and inorganic crystallization processes1,2,3,4,5. A two-step nucleation theory has thus been proposed, in which initial low-density, solvated amorphous aggregates subsequently densify, leading to nucleation4,6,7. This view differs from classical nucleation theory, which implies that crystalline nuclei forming in solution have the same density and structure as does the final crystalline state1. A protein crystallization mechanism involving this classical pathway has recently been observed directly8. However, a molecular mechanism of nonclassical protein crystallization9,10,11,12,13,14,15 has not been established9,11,14. To determine the nature of the amorphous precursors and whether crystallization takes place within them (and if so, how order develops at the molecular level), three-dimensional (3D) molecular-level imaging of a crystallization process is required. Here we report cryogenic scanning transmission microscopy tomography of ferritin aggregates at various stages of crystallization, followed by 3D reconstruction using simultaneous iterative reconstruction techniques to provide a 3D picture of crystallization with molecular resolution. As crystalline order gradually increased in the studied aggregates, they exhibited an increase in both order and density from their surface towards their interior. We observed no highly ordered small structures typical of a classical nucleation process, and occasionally we observed several ordered domains emerging within one amorphous aggregate, a phenomenon not predicted by either classical or two-step nucleation theories. Our molecular-level analysis hints at desolvation as the driver of the continuous order-evolution mechanism, a view that goes beyond current nucleation models, yet is consistent with a broad spectrum of protein crystallization mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that supports the findings of this study are available from the corresponding authors upon request.
Code availability
The program code for the database implemented orientation order parameter analysis performed in this manuscript is available from https://github.com/LotharHouben/diOpa.
References
De Yoreo, J. J. D. et al. Crystallization by particle attachment in synthetic, biogenic, and geologic environments. Science 349, aaa6760 (2015).
Gebauer, D., Kellermeier, M., Gale, J. D., Bergstrom, L. & Colfen, H. Pre-nucleation clusters as solute precursors in crystallisation. Chem. Soc. Rev. 43, 2348–2371 (2014).
Sear, R. P. The non-classical nucleation of crystals: microscopic mechanisms and applications to molecular crystals, ice and calcium carbonate. Int. Mater. Rev. 57, 328–356 (2012).
Vekilov, P. G. Nucleation. Cryst. Growth Des. 10, 5007–5019 (2010).
Anwar, J. & Zahn, D. Uncovering molecular processes in crystal nucleation and growth by using molecular simulation. Angew. Chem. Int. Ed. 50, 1996–2013 (2011).
ten Wolde, P. R. & Frenkel, D. Enhancement of protein crystal nucleation by critical density fluctuations. Science 277, 1975–1978 (1997).
Erdemir, D., Lee, A. Y. & Myerson, A. S. Nucleation of crystals from solution: classical and two-step models. Acc. Chem. Res. 42, 621–629 (2009).
Van Driessche, A. E. S. et al. Molecular nucleation mechanisms and control strategies for crystal polymorph selection. Nature 556, 89–94 (2018).
Sleutel, M. & Van Driessche, A. E. S. Nucleation of protein crystals—a nanoscopic perspective. Nanoscale 10, 12256–12267 (2018).
Zhang, F. J. Nonclassical nucleation pathways in protein crystallization. J. Phys. Condens. Mat. 29, 443002 (2017).
Vekilov, P. G. Nucleation of protein crystals. Prog. Cryst. Growth 62, 136–154 (2016).
Yamazaki, T. et al. Two types of amorphous protein particles facilitate crystal nucleation. Proc. Natl Acad. Sci. USA 114, 2154–2159 (2017).
Sauter, A. et al. Real-time observation of nonclassical protein crystallization kinetics. J. Am. Chem. Soc. 137, 1485–1491 (2015).
Vekilov, P. G. & Vorontsova, M. A. Nucleation precursors in protein crystallisation. Acta Crystallogr. F 70, 271–282 (2014).
Sleutel, M. & Van Driessche, A. E. S. Role of clusters in nonclassical nucleation and growth of protein crystals. Proc. Natl Acad. Sci. USA 111, E546–E553 (2014).
Crystallization Recipes: Ferritin https://www1.rigaku.com/ja/node/2746#Ferritin (Rigaku Corporation, 2020).
Wolf, S. G., Houben, L. & Elbaum, M. Cryo-scanning transmission electron tomography of vitrified cells. Nat. Methods 11, 423–428 (2014).
Granier, T., Gallois, B., Dautant, A., Langlois d’Estaintot, B. & Précigoux, G. Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin. Acta Crystallogr. D 53, 580–587 (1997).
Janssens, K. G. F. et al. Computing the mobility of grain boundaries. Nat. Mater. 5, 124–127 (2006).
Yau, S. T. & Vekilov, P. G. Quasi-planar nucleus structure in apoferritin crystallization. Nature 406, 494–497 (2000).
Steinhardt, P. J., Nelson, D. R. & Ronchetti, M. Bond-orientational order in liquids and glasses. Phys. Rev. B 28, 784–805 (1983).
Gasser, U., Ziese, F. & Maret, G. Characterization of local structures with bond-order parameters and graphs of the nearest neighbors, a comparison. Eur. Phys. J. Spec. Top. 223, 455–467 (2014).
Lechner, W. & Dellago, C. Accurate determination of crystal structures based on averaged local bond order parameters. J. Chem. Phys. 129, 114707 (2008).
Acknowledgements
This work was supported by the Helen and Martin Kimmel Center for Molecular Design. We thank S. Albeck, M. Peretz and J. Jacobovitch for assistance with GPC experiments, I. Biran for help acquiring cryo-STEM tomograms, and B. Palmer and R. Diskin for discussions. The electron microscopy studies were partially supported by the Irving and Cherna Moskowitz Center for Nano and BioNano Imaging (Weizmann Institute of Science). S.G.W. acknowledges support for cryo-STEM tomography studies from the Israel Science Foundation (grant number 1285/14).
Author information
Authors and Affiliations
Contributions
B.R., L.H. and H.W. conceived the project. H.W. performed the crystallization experiments and the 2D cryo-TEM imaging; S.G.W., H.W. and L.H. performed the cryo-STEM tomography imaging; and L.H performed the cryo-STEM tomography data analysis. B.R. and L.H. wrote the manuscript. All authors contributed to the discussion and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Mike Sleutel, Peter Vekilov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Description, Supplementary Figures and references related to crystallization procedures, Cryo-(S)TEM experiments, and order analysis methods.
Video 1
An animated representation of the procedure for the identification of ferritin coordinates from cryo-STEM data. The movie shows in sequence a tilt series of annular dark field images of a ferritin aggregate after image alignment, tomogram reconstruction slices in the viewing direction from the top of the cryo sample, the identification of peak signals associated with iron-rich cores of ferritin monomers in the two-dimensional reconstruction slices, and the remaining ferritin coordinates in the 3D tomogram volume after refinement that eliminates the peak overlap in the third dimension.
Video 2
An animated representation of a highly ordered ferritin aggregate in a cryo-STEM tilt series of bright-field images and the tomogram reconstruction slices in the viewing direction from the top. SI Video 2 exemplifies the accurate reproduction of the crystalline structure with monomer resolution.
Rights and permissions
About this article
Cite this article
Houben, L., Weissman, H., Wolf, S.G. et al. A mechanism of ferritin crystallization revealed by cryo-STEM tomography. Nature 579, 540–543 (2020). https://doi.org/10.1038/s41586-020-2104-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2104-4
This article is cited by
-
Non-classical crystallization in soft and organic materials
Nature Reviews Materials (2024)
-
Onion-like multicolor thermally activated delayed fluorescent carbon quantum dots for efficient electroluminescent light-emitting diodes
Nature Communications (2024)
-
Protein nanocondensates: the next frontier
Biophysical Reviews (2023)
-
Atomic-scale observation of non-classical nucleation-mediated phase transformation in a titanium alloy
Nature Materials (2022)
-
Nucleation of protein mesocrystals via oriented attachment
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.