Abstract
Current programmable nuclease-based methods (for example, CRISPR–Cas9) for the precise correction of a disease-causing genetic mutation harness the homology-directed repair pathway. However, this repair process requires the co-delivery of an exogenous DNA donor to recode the sequence and can be inefficient in many cell types. Here we show that disease-causing frameshift mutations that result from microduplications can be efficiently reverted to the wild-type sequence simply by generating a DNA double-stranded break near the centre of the duplication. We demonstrate this in patient-derived cell lines for two diseases: limb-girdle muscular dystrophy type 2G (LGMD2G)1 and Hermansky–Pudlak syndrome type 1 (HPS1)2. Clonal analysis of inducible pluripotent stem (iPS) cells from the LGMD2G cell line, which contains a mutation in TCAP, treated with the Streptococcus pyogenes Cas9 (SpCas9) nuclease revealed that about 80% contained at least one wild-type TCAP allele; this correction also restored TCAP expression in LGMD2G iPS cell-derived myotubes. SpCas9 also efficiently corrected the genotype of an HPS1 patient-derived B-lymphoblastoid cell line. Inhibition of polyADP-ribose polymerase 1 (PARP-1) suppressed the nuclease-mediated collapse of the microduplication to the wild-type sequence, confirming that precise correction is mediated by the microhomology-mediated end joining (MMEJ) pathway. Analysis of editing by SpCas9 and Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a) at non-pathogenic 4–36-base-pair microduplications within the genome indicates that the correction strategy is broadly applicable to a wide range of microduplication lengths and can be initiated by a variety of nucleases. The simplicity, reliability and efficacy of this MMEJ-based therapeutic strategy should permit the development of nuclease-based gene correction therapies for a variety of diseases that are associated with microduplications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw data associated with Extended Data Fig. 10 are reported in Supplementary Table 3. Raw script used for retrieving microduplication data listed in Supplementary Table 3 will be available upon request. Raw Illumina sequencing reads and PacBio data for this study have been deposited in the National Center for Biotechnology Information Short Read Archive under bioproject ID PRJNA517630.
Code availability
Data analysis used a combination of publicly available software and custom code, as detailed in the Methods. Custom Python (CRESA-lpp.py) and R (indel_background_filtering.R) scripts used in the Illumina data analysis and the shell script (Tcap_pacbio_analysis.sh) used for the analysis of the PacBio data are hosted on GitHub (https://github.com/locusliu/PCR_Amplicon_target_deep_seq). Scripts for the bioinformatic analysis of pathogenic microduplications are hosted at https://rambutan.umassmed.edu/duplications/.
References
Moreira, E. S. et al. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat. Genet. 24, 163–166 (2000).
El-Chemaly, S. & Young, L. R. Hermansky–Pudlak syndrome. Clin. Chest Med. 37, 505–511 (2016).
Sfeir, A. & Symington, L. S. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714 (2015).
Bae, S., Kweon, J., Kim, H. S. & Kim, J.-S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–706 (2014).
Kim, S.-I. et al. Microhomology-assisted scarless genome editing in human iPSCs. Nat. Commun. 9, 939 (2018).
Hisano, Y. et al. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 5, 8841 (2015).
Sakuma, T., Nakade, S., Sakane, Y., Suzuki, K. T. & Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR–Cas9 with the PITCh systems. Nat. Protoc. 11, 118–133 (2016).
Bertz, M., Wilmanns, M. & Rief, M. The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc. Natl Acad. Sci. USA 106, 13307–13310 (2009).
Nigro, V. & Savarese, M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 33, 1–12 (2014).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Caron, L. et al. A human pluripotent stem cell model of facioscapulohumeral muscular dystrophy-affected skeletal muscles. Stem Cells Transl. Med. 5, 1145–1161 (2016).
Oh, J. et al. Positional cloning of a gene for Hermansky–Pudlak syndrome, a disorder of cytoplasmic organelles. Nat. Genet. 14, 300–306 (1996).
Richmond, B. et al. Melanocytes derived from patients with Hermansky–Pudlak syndrome types 1, 2, and 3 have distinct defects in cargo trafficking. J. Invest. Dermatol. 124, 420–427 (2005).
Brantly, M. et al. Pulmonary function and high-resolution CT findings in patients with an inherited form of pulmonary fibrosis, Hermansky–Pudlak syndrome, due to mutations in HPS-1. Chest 117, 129–136 (2000).
Bolukbasi, M. F. et al. Orthogonal Cas9–Cas9 chimeras provide a versatile platform for genome editing. Nat. Commun. 9, 4856 (2018).
Sharma, S. et al. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 6, e1697 (2015).
Wang, M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006).
Dutta, A. et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 45, 2585–2599 (2017).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771 (2015).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726 (2019).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Bolukbasi, M. F. et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat. Methods 12, 1150–1156 (2015).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Rittié, L. & Fisher, G. J. Isolation and culture of skin fibroblasts. Methods Mol. Med. 117, 83–98 (2005).
Stadler, G. et al. Establishment of clonal myogenic cell lines from severely affected dystrophic muscles — CDK4 maintains the myogenic population. Skelet. Muscle 1, 12 (2011).
Kearns, N. A. et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141, 219–223 (2014).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014).
Blankenberg, D. et al. Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785 (2010).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Tan, A., Abecasis, G. R. & Kang, H. M. Unified representation of genetic variants. Bioinformatics 31, 2202–2204 (2015).
Obenchain, V. et al. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30, 2076–2078 (2014).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Li, H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–1844 (2012).
Wu, Y. et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. https://doi.org/10.1038/s41591-019-0401-y (2019).
Liu, P. et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz184 (2019).
Acknowledgements
We thank E. Kittler and the UMass Medical School Deep Sequencing Core for sequencing; L. Hayward, L. Qin and D. McKenna-Yasek for coordinating patient enrolment and acquiring patient skin biopsies; Z. Matijasevic for generating LGMD2G iPS cell lines; and the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. This work was supported in part by the National Institutes of Health (R01DK098252, R01HL131471 and R01NS088689 (C.M.); R01AI117839, R01GM115911, R01HL093766 and U01HG007910 (S.A.W.); U54HD0060848 (C.P.E.); a SPARK award through UL1-TR001453) and the Worcester Foundation for Biomedical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Reviewer information
Nature thanks Randall Platt and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.I. and S.S. performed and analysed the B-LCL and HEK293T editing experiments. D.G., J.C.J.C. and K.D. performed the iPS cell and myoblast editing experiments. S.I. and S.S. analysed the iPS cell and myoblast editing experiments. D.G. and J.C.J.C. generated the iPS cells and myoblast cell lines. D.G. and K.D. performed and analysed the flow cytometry and western blot data for detection of telethonin. B.P.R., P.L. and K.L. designed and purified the SpCas9 and LbCas12a proteins. P.L. and O.D.K. performed the bioinformatic analysis. P.L. analysed the deep sequencing data. C.M. and M.Z. contributed expertise to the HPS1 cell line editing and characterization. C.M., O.D.K., C.P.E. and S.A.W. directed the research. S.I., S.S., C.P.E. and S.A.W. wrote the manuscript with input from all of the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors have filed patent applications related to genome engineering technologies. S.I., S.S., D.G., J.C.J.C., C.M., O.D.K., C.P.E. and S.A.W. have filed a patent application (62/667201) on this work. C.M. is a paid consultant for and with equity in Apic Bio. Apic Bio was not involved in the funding or any other aspects of this study. The authors have no other competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
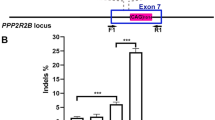
Extended Data Fig. 1 Indel populations resulting from SpCas9 editing at the TCAP locus.
a, Indel percentages resulting from SpCas9 RNP treatment in patient-derived iPS cells homozygous for the 8-bp microduplication or in wild-type iPS cells. Mean ± s.e.m. from three biological replicates. b, Breakdown of indel classes resulting from SpCas9 treatment of myoblasts derived from patient-derived LGMD2G iPS cells. Mean ± s.e.m. from three biological replicates. c, Sequence alignment of the edited alleles resulting from SpCas9 RNP treatment of LGMD2G iPS cells. Red and blue text indicates DNA repeats that constitute the microduplication, and collapse is indicated by half red and half blue text. Dashes indicate deleted bases and purple text indicates inserted bases. Data are from one biological replicate out of three independent biological replicates. d, Sequence alignment of the edited alleles resulting from SpCas9 RNP treatment of myoblasts derived from patient-derived LGMD2G iPS cells. Data are from one biological replicate out of three independent biological replicates.
Extended Data Fig. 2 PacBio long-read sequencing analysis for SpCas9-edited LGMD2G iPS cells at the TCAP locus.
a, Percentage of gene modification observed from PacBio sequencing (one replicate from Fig. 1c out of three biological replicates). Green, alleles containing the 8-bp deletion; grey, other small indels(≤100 bp); blue, large insertions (0.14%, not visible on the graph); maroon, large deletions (>100 bp). b, IGV graphs depicting representative reads obtained for unedited (top) and edited (bottom) LGMD2G iPS cells, spanning a genomic region of about 2,035 bp surrounding the TCAP target site. Red caret indicates the 8-bp deletion site. Data represent one replicate out of three independent biological replicates.
Extended Data Fig. 3 PacBio long-read sequencing analysis of SpCas9-edited LGMD2G iPS cells clones and a complex colony at the TCAP locus.
IGV graphs depicting representative reads obtained for clonal isolates of edited LGMD2G iPS cells (Fig. 1d), spanning a genomic region of about 2,035 bp surrounding the TCAP target site. The genotype of the clones (deduced by Illumina deep sequencing) is indicated beside an enlargement of the TCAP target region within the PacBio data. The sequences of the two alleles (listed above the IGV plot) obtained from sequencing are shown with repeats in red and blue. Alleles that reverted to wild-type as a result of collapse of microduplication are half red/half blue. Bottom, IGV plot for one complex iPS cell colony that appears to have been nucleated by more than one cell, with large deletions present in the genome (sizes indicated).
Extended Data Fig. 4 Detection of telethonin expression by flow cytometry in patient-derived cells treated with SpCas9.
a, Contour plots from a representative flow cytometry assay to detect telethonin expression in healthy control cells (TCAP+/+), patient cells (TCAP−/−), and SpCas9-treated homozygous and heterozygous iPS clone-derived myoblasts differentiated for 10 days in culture. Plots are representative of three independent replicates. b, Histograms from a representative flow cytometry assay to detect telethonin expression. Left, overlay of anti-telethonin antibody staining for four representative samples for different TCAP genotypes. Right, comparison between patient cells and healthy control cells, and SpCas9-treated homozygous and heterozygous iPS clone-derived myoblasts differentiated for 10 days in culture. Histograms are representative of three independent replicates. c, Cells were selected by removing cell debris first as shown by gate P1, and then single cells were selected from P1 by removing clustered cells as shown by gate P2. The cells in gate P2 were used for flow analysis. Plots are representative of one biological replicate. d, Average percentage of telethonin-expressing cells from two technical replicates of three biological replicates. Error bars indicate s.e.m (n = 6) and circles represent individual data points. P values (P = 0.33 for patient versus heterozygous and *P = 0.04 for patient versus homozygous clones) were calculated by two-sided Student’s t-test (Supplementary Table 9). ns, not significant. e, Western blot showing validation of anti-telethonin antibody (Santa Cruz Biotechnology). Human muscle lysate and lysate from HEK293T cells transfected with haemagglutinin-tagged telethonin expression construct were separated on an SDS 4–12% acrylamide gradient gel and the resulting blot was probed with anti-telethonin antibody. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 Standard curve generated with genomic DNA of wild-type and HPS1 mutant B-LCLs from UMI-based Illumina deep sequencing.
Genomic DNA from wild-type cells and HPS1 cells homozygous for the 16-bp microduplication were mixed at different ratios (x axis). These mixed DNAs were used for the construction of a UMI-based Illumina library to determine the ratio of the alleles through deep sequencing (y axis). These data are fitted to a regression line with the R2 value reported. n = 1 biological replicate.
Extended Data Fig. 6 Indel spectrum generated by SpCas9 editing at the HPS1 locus in HPS1 B-LCL cells.
Indel spectra of SpCas9 nuclease cells treated with different sgRNAs determined by UMI-based Illumina deep sequencing. a, Target site 1. b, Target site 2. c, Target site 3. d, Target site 4. e, Target site 5. f, Target site 6. Red bar indicates 16-bp deletion that corresponds to the deletion of one of the microduplication repeats. Data show indel spectra from one representative biological replicate out of three independent biological replicates.
Extended Data Fig. 7 Effect of rucaparib on the profile of microhomology-mediated deletion products at AAVS1 locus in patient-derived HPS1 B-LCL cells.
a, Schematic of two prominent DNA DSB repair pathways. A DSB can be repaired through various pathways that produce different DNA sequence end products. The NHEJ pathway is the dominant DSB repair pathway in most cells. The MMEJ pathway uses end-resection to discover small homologies on each side of the break that can be used to template the fusion of the broken ends. PARP-1 regulates DSB flux through the MMEJ pathway. Treatment of cells with rucaparib—an inhibitor of PARP-1—attenuates DSB flux down the MMEJ repair pathway. b, Percentage of microhomology-mediated deletions (green) and total indels (blue) resulting from SpCas9 treatment of cells in the presence of 0,10 and 20 μM rucaparib. Bars show mean and dots show individual data points from three biological replicates based on UMI-based Illumina deep sequencing. c, Percentage of 1-bp insertions (purple), microhomology mediated deletions (green) and other deletions (grey) produced by SpCas9 RNP with a sgRNA targeting the AAVS1 locus with the addition of increasing amounts of rucaparib. Mean ± s.e.m. from three biological replicates based on UMI-based Illumina deep sequencing. d, Percentage of microhomology-mediated deletions out of total indels in cells treated with SpCas9 in the presence of rucaparib. Mean ± s.e.m., dots represent individual data points from three biological replicates. P values determined using two-tailed unpaired t-test (Supplementary Table 9). ***P = 0.0004, ****P = 6.5 × 10−7. e, Left, alignment of allele sequences obtained from deep sequencing analysis from samples treated with SpCas9 RNP in the presence of different rucaparib concentrations. Microhomologies present at the AAVS1 locus are shown in by red, green and blue. Microhomology-mediated deletion is indicated by two-toned text. Magenta carets indicate site of DSB created by SpCas9. Inserted bases (ins) are shown in purple, deleted bases (del) are shown as black dashes. Right, heat map depicting the percentage of alleles generated after SpCas9 treatment of cells in the presence of different concentrations of rucaparib (0, 10 or 20 μM). The blue colour gradient scale indicates the percentage of occurrence of that sequence. Heat map represents mean values from a total of three independent biological replicates.
Extended Data Fig. 8 Editing with SpCas9 and LbCas12a at endogenous microduplications.
a, Percentage of microhomology-mediated deletions out of total indels at endogenous sites in cells treated with SpCas9 and LbCas12a. Mean ± s.e.m., dots represent individual data points from three biological replicates. b, Schematic of endogenous site containing a 24-bp microduplication for SpCas9 target sites 1–3. The 24-bp microduplication repeats are shown in bold red and blue. The PAM sequence is outlined in magenta and the protospacer sequence is underlined. Magenta carets indicate the site of DSB. c, Percentage of alleles with 24-bp deletion (green) and total indels (blue) for all three guides from TIDE analysis. Guide 3 produces primarily 23-bp deletions, but not 24-bp deletions, probably because it recuts the collapsed DNA sequence. Bars shows the mean from n = 3 biological repeats, individual data points are represented by dots. d, Proportion of the 24-bp deletion out of total indels as individual data points (dots), with mean ± s.e.m. n = 3 biological repeats. e, Schematic of endogenous site containing a 27-bp microduplication for SpCas9 target sites 1 and 2. f, Percentage of alleles with 27-bp deletion (green) and total indels (blue) for both guides from UMI-based Illumina deep sequencing. Bars show the mean from n = 3 biological repeats, individual data points are represented by dots. g, Proportion of the 27-bp deletion out of total indels as individual data points (dots) with mean ± s.e.m. n = 3 biological replicates.
Extended Data Fig. 9 Bioinformatics pipeline for identification of disease alleles.
Schematic shows the bioinformatics pipeline used to identify all microduplications amendable to efficient MMEJ-mediated collapse from the ‘coding’ regions (exome_calling_regions.v1; mainly exons plus 50 flanking bases) in the gnomAD genome and exome databases (version 2.0.2). Insertion variants observed in both databases were used for analysis (variants occurring in both databases were counted once). Insertions that do not add a repeat unit to an existing tandem repeat and are not themselves a perfect repeat were filtered to constrain only duplications that spanned 2–40 bp in length and are amendable to CRISPR–Cas9 targeting. This dataset was then cross-referenced against the ClinVar database (clinvar_20180225.vcf) to apply further filters for variants reported as pathogenic, which ultimately yielded 143 likely disease-causing microduplications.
Extended Data Fig. 10 Pathogenic microduplications and their prevalence in human populations.
a, Number of insertion variants of length >1 bp that are annotated as pathogenic or pathogenic/likely pathogenic in ClinVar. Variants are binned by length, with all those of length 40 bp or greater combined. The insertions (grey) are stratified into progressively finer categories: duplications (red); ‘simple’ duplications (described in text, orange); and the subset of these observed at least once in gnomAD exome/genome databases (green). b, Number of insertion variants of length >1 bp that are observed at least once in the ‘coding’ regions of the gnomAD exome/genome databases. As above, insertions (grey) are stratified into progressively finer categories: duplications (red); ‘simple’ duplications (orange); the subset of these listed in ClinVar (cyan); and the subset annotated as Pathogenic or Pathogenic/likely pathogenic in ClinVar (green). Cyan and green bars are not visible at this resolution.
Supplementary information
Supplementary Information
This file contains Supplementary Discussion, legends for Supplementary Tables 1-9, and Supplementary Figure 1. Supplementary Discussion: 143 disease alleles associated with microduplications identified in this study does not reflect the upper limit of prevalence of pathogenic microduplications in the human population. Supplementary Figure: Western blot showing validation of anti-telethonin antibody (uncropped gel source data for Extended Data Figure 4).
Supplementary Tables
This file contains Supplementary Tables 1–9.
Source data
Rights and permissions
About this article
Cite this article
Iyer, S., Suresh, S., Guo, D. et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565 (2019). https://doi.org/10.1038/s41586-019-1076-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1076-8
This article is cited by
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Molecular Biomedicine (2023)
-
Prime editing for precise and highly versatile genome manipulation
Nature Reviews Genetics (2023)
-
Functional restoration of mouse Nf1 nonsense alleles in differentiated cultured neurons
Journal of Human Genetics (2022)
-
Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining
Nature Reviews Molecular Cell Biology (2022)
-
PARP inhibitors in gastric cancer: beacon of hope
Journal of Experimental & Clinical Cancer Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.