Abstract
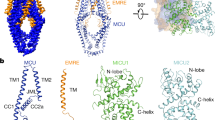
The mitochondrial calcium uniporter (MCU) is a highly selective calcium channel localized to the inner mitochondrial membrane. Here, we describe the structure of an MCU orthologue from the fungus Neosartorya fischeri (NfMCU) determined to 3.8 Å resolution by phase-plate cryo-electron microscopy. The channel is a homotetramer with two-fold symmetry in its amino-terminal domain (NTD) that adopts a similar structure to that of human MCU. The NTD assembles as a dimer of dimers to form a tetrameric ring that connects to the transmembrane domain through an elongated coiled-coil domain. The ion-conducting pore domain maintains four-fold symmetry, with the selectivity filter positioned at the start of the pore-forming TM2 helix. The aspartate and glutamate sidechains of the conserved DIME motif are oriented towards the central axis and separated by one helical turn. The structure of NfMCU offers insights into channel assembly, selective calcium permeation, and inhibitor binding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
14 August 2018
In this Article, ref. 15 has been replaced and references 32 to 50 have been renumbered online.
References
Kamer, K. J. & Mootha, V. K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553 (2015).
Berridge, M. J., Bootman, M. D. & Roderick, H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 (2003).
Kirichok, Y., Krapivinsky, G. & Clapham, D. E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427, 360–364 (2004).
Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011).
De Stefani, D., Raffaello, A., Teardo, E., Szabò, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Sancak, Y. et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382 (2013).
Plovanich, M. et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8, e55785 (2013).
Perocchi, F. et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467, 291–296 (2010).
Tsai, M.-F. et al. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife 5, e15545 (2016).
Kovács-Bogdán, E. et al. Reconstitution of the mitochondrial calcium uniporter in yeast. Proc. Natl Acad. Sci. USA 111, 8985–8990 (2014).
Carafoli, E. & Lehninger, A. L. A survey of the interaction of calcium ions with mitochondria from different tissues and species. Biochem. J. 122, 681–690 (1971).
Bick, A. G., Calvo, S. E. & Mootha, V. K. Evolutionary diversity of the mitochondrial calcium uniporter. Science 336, 886 (2012).
Song, J., Liu, X., Zhai, P., Huang, J. & Lu, L. A putative mitochondrial calcium uniporter in A. fumigatus contributes to mitochondrial Ca2+ homeostasis and stress responses. Fungal Genet. Biol. 94, 15–22 (2016).
Lee, Y. et al. Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter. EMBO Rep. 16, 1318–1333 (2015).
Lee, S. K. et al. Structural insights into mitochondrial calcium uniporter regulation by divalent cations. Cell Chem. Biol. 23, 1157–1169 (2016).
Oxenoid, K. et al. Architecture of the mitochondrial calcium uniporter. Nature 533, 269–273 (2016).
Wang, L. et al. Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake. EMBO J. 33, 594–604 (2014).
Logan, C. V. et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 46, 188–193 (2014).
Gunter, T. E. & Pfeiffer, D. R. Mechanisms by which mitochondria transport calcium. Am. J. Physiol. 258, C755–C786 (1990).
Frauenfeld, J. et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods 13, 345–351 (2016).
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998).
Biedermannova, L., E Riley, K., Berka, K., Hobza, P. & Vondrasek, J. Another role of proline: stabilization interactions in proteins and protein complexes concerning proline and tryptophane. Phys. Chem. Chem. Phys. 10, 6350–6359 (2008).
Zondlo, N. J. Aromatic-proline interactions: electronically tunable CH/π interactions. Acc. Chem. Res. 46, 1039–1049 (2013).
Mallilankaraman, K. et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644 (2012).
Kamer, K. J. & Mootha, V. K. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 15, 299–307 (2014).
Patron, M. et al. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell 53, 726–737 (2014).
Liu, J. C. et al. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload. Cell Reports 16, 1561–1573 (2016).
Almers, W. & McCleskey, E. W. Non-selective conductance in calcium channels of frog muscle: calcium selectivity in a single-file pore. J. Physiol. (Lond.) 353, 585–608 (1984).
Hess, P. & Tsien, R. W. Mechanism of ion permeation through calcium channels. Nature 309, 453–456 (1984).
Heinemann, S. H., Terlau, H., Stühmer, W., Imoto, K. & Numa, S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 356, 441–443 (1992).
Cao, C., Wang, S., Cui, T., Su, X.-C. & Chou, J. J. Ion and inhibitor binding of the double-ring ion selectivity filter of the mitochondrial calcium uniporter. Proc. Natl Acad. Sci. USA 114, E2846–E2851 (2017).
Qing, G. et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882 (2004).
Danev, R., Tegunov, D. & Baumeister, W. Using the Volta phase plate with defocus for cryo-EM single particle analysis. eLife 6, e23006 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
DiMaio, F., Zhang, J., Chiu, W. & Baker, D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 22, 865–868 (2013).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Schrödinger, L. The PyMOL Molecular Graphics System, Version 1.8. (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Yaffe, M. P. Analysis of mitochondrial function and assembly. Methods Enzymol. 194, 627–643 (1991).
Lo, C. J., Leake, M. C., Pilizota, T. & Berry, R. M. Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys. J. 93, 294–302 (2007).
Heginbotham, L., Kolmakova-Partensky, L. & Miller, C. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998).
Pei, J., Kim, B. H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Acknowledgements
We thank J. Guo and Q. Chen for technical assistance and manuscript preparation, and J. Frauenfeld (Salipro Biotech) for the recombinant saposin construct. Negative-stained EM data acquisition and sample optimization for cryo-EM were performed at the University of California San Francisco; we thank M. Braunfeld for supervision of the microscopes. Phase plate cryo-EM data were collected at the University of Texas Southwestern Medical Center Cryo-EM Facility, which is funded by the CPRIT Core Facility Support Award RP170644; we thank D. Nicastro and Z. Chen for technical support and facility access. This work was supported in part by the Howard Hughes Medical Institute (to Y.J., V.K.M. and Y.C.), the National Institutes of Health (grant GM079179 to Y.J.; grants R01GM098672, S10OD020054, and S10OD021741 to Y.C.), the Welch Foundation (Grant I-1578 to Y.J.), the Cancer Prevention and Research Initiative of Texas (to X.B.) and the Virginia Murchison Linthicum Scholar in Medical Research fund (to X.B.).
Reviewer information
Nature thanks M. Prakriya, D. Stokes and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
N.X.N., V.K.M. and Y.J. conceived the study. N.X.N., J.-P.A., Y.J., Y.C. and X.B. designed the experiments and analysed the data. N.X.N., J.-P.A., C.L., Y.Y., W.Z., and X.B. generated key materials and executed the experiments. N.X.N. and Y.J. wrote the manuscript with input from J.-P.A., Y.C., V.K.M. and X.B. Y.J. supervised the project and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Biochemical analysis of NfMCU.
a, Representative gel filtration profile of full-length (black trace) versus truncated NfMCU used for cryo-EM studies (blue trace). b, Functional activity of full-length (FL) and truncated NfMCU (EM); data shown represent mean ± s.e.m. (n = 3 independent experiments). Cell lysates were analysed by immunoblotting using anti-His antibody to detect expression of NfMCU; RpoA was used as loading control. c, Detergent-solubilized NfMCU (control) separates as a single band at ~46 kDa on Coomassie-stained SDS–PAGE and crosslinks as tetramers when treated with chemical crosslinkers of varying spacer arm length. DSG, disuccinimidyl glutarate; DSS, disuccinimidyl suberate; BS3, bis(sulfosuccinimidyl) suberate; BSPEG5, PEGylated bis(sulfosuccinimidyl)suberate. See Supplementary Fig. 1 for gel source data. The experiments shown in a and b were repeated at least three times independently with similar results. The crosslinking experiment shown in c was performed once.
Extended Data Fig. 2 Functional characterization of NfMCU.
a, Calcium uptake in E. coli expressing empty vector (black trace) or NfMCU (blue trace), demonstrating that heterologously expressed NfMCU can confer uniporter function to bacteria. Arrows indicate addition of 20 µM CaCl2 to the reaction solution. b, Concentration-dependent calcium uptake in E. coli expressing empty vector (black trace) or NfMCU (blue trace). Each point represents the rate of calcium uptake normalized to the uptake rate at 90 µM CaCl2. c, Concentration-dependent inhibition of NfMCU by GdCl3 in E. coli expressing NfMCU. Each point represents the rate of calcium uptake normalized to the uptake rate without GdCl3 in the reaction. d, Concentration-dependent inhibition of Ca2+ uptake in E. coli expressing NfMCU by the MCU-specific inhibitor Ru360. Each point represents the rate of calcium uptake normalized to the uptake rate without Ru360 in the reaction. The Ru360 concentration required to reduce the Ca2+ uptake rate by half (IC50) is calculated to be 0.5 μM. e, In light of the contradiction of the oligomeric state between NfMCU and cMCU-∆NTD, we also designed an in vitro functional assay by reconstituting our purified tetrameric channel into liposomes. Unable to obtain currents of NfMCU by electrophysiology—probably owing to the channel’s small conductance—we performed a radioactive 45Ca flux assay on proteoliposomes loaded with 150 mM KCl. Shown here is the time-dependent 45Ca2+ uptake by empty liposomes or NfMCU proteoliposomes incubated with 45Ca2+ in a reaction buffer containing 150 mM KCl (red points) or 150 mM NMDG (blue points). The result demonstrates that in a reaction solution containing 150 mM NMDG and the K+-selective ionophore valinomycin (to generate an electrical driving force), NfMCU proteoliposomes exhibited time-dependent 45Ca2+ uptake while the same proteoliposomes in a reaction solution containing valinomycin and 150 mM KCl (to eliminate electrical driving force) accumulated 45Ca2+ only slightly better than empty liposomes, recapitulating the voltage-dependent Ca2+ uptake property of the MCU. f, Concentration-dependent inhibition of NfMCU proteoliposomes by Ru360, demonstrating that Ru360 also blocks NfMCU 45Ca2+ uptake in a concentration-dependent manner up to 50%, which is expected given an equal distribution of two channel orientations in the liposomes. Each point represents radioactivity measured after 30 min reaction normalized to sample without Ru360. All data in a–f are shown as mean ± s.e.m. (n = 3 independent experiments).
Extended Data Fig. 3 Cryo-EM analysis of NfMCU.
a, Representative phase-plate electron micrograph of the NfMCU saposin complex and 2,470 micrographs were used for structure determination. b, Representative 2D class averages of the NfMCU saposin complex. c, Flowchart of data processing to obtain the 3.8 Å cryo-EM map of the NfMCU saposin complex. Details can be found in the Image processing section of the Methods. d, Local resolution of NfMCU estimated with RELION2.0. e, FSC curves for the refined model versus the summed 3.8 Å map (black curve), the refined model versus half map 1 (red curve), and the refined model versus half map 2 that was not used for refinement (green curve). f, The gold-standard FSC curve for the cryo-EM map.
Extended Data Fig. 4 EM map of NfMCU.
a, Representative regions of the EM map of NfMCU highlighting key structural features at the NTD (α1, α2, β2– β3), the coiled-coil domain, and transmembrane domain (TM1 and TM2). Protomers 1 and 2 are shown in yellow and orange, respectively. b, Stereo view of the EM map carved around the TM2 filter region of NfMCU.
Extended Data Fig. 5 Cryo-EM structure of NfMCU in complex with saposin.
a, Cryo-EM map of NfMCU in complex with saposin (shown alternating between purple and pink). The map is low-pass filtered to 5 Å to allow better visualization of the density from the saposin molecules. Six saposin molecules, each oriented at a 45° angle to the vertical axis, wrap around NfMCU stabilized in lipid. b, Ribbon diagram of NfMCU in complex with saposin. Approximately 79–81 residues of the 81-residue saposin molecule were modelled as a poly-alanine chain into the helix–turn–helix EM map density for saposin.
Extended Data Fig. 6 Sequence alignment of MCU orthologues.
The sequences were aligned using PROMALS3D50 and numbered according to NfMCU. Secondary structures shown above the sequences are based on the cryo-EM structure of NfMCU. Loops are denoted by solid black lines; disordered regions not observed in the cryo-EM structure and truncations made to NfMCU are indicated by dashed lines. The first 75 residues in NfMCU (grey arrow) are predicted to be the mitochondrial targeting sequence (MTS). NCBI accession number for the sequences are: HsMCU (human MCU): NP_612366.1, MmMCU (mouse MCU): XP_006513531.1, DdMCU (D. discoideum MCU): XP_637750.1, CeMCU (C. elegans MCU): NP_500892.1, NcMCU (Neurospora crassa MCU): XP_959658.1, NfMCU (Neosartorya fischeri MCU): XP_001266985.1.
Extended Data Fig. 7 Structural comparison between MCU domains.
a, Overall structure comparison between tetrameric NfMCU and pentameric cMCU-∆NTD (PDB: 5ID3). Each subunit is individually coloured. b, Structure comparison between a single subunit of the pore domain of NfMCU and cMCU-∆NTD. Dashed boxes indicate major differences between the two structures. Structural features in cMCU-∆NTD are labelled as in the original structure16. IJMH, inner juxtamembrane helix; OJMH, outer juxtamembrane helix; L1, loop 1; L2, loop 2; JML, juxtamembrane loop; CC1, coiled-coil 1; CC2, coiled-coil 2. c, Structural comparison between the NTDs of NfMCU and HsMCU (PDB: 4XTB). Dashed box indicates the major difference between the two structures. d, Atomic interactions between two NTD subunits in the crystal structure of HsMCU NTD, equivalent to interface I identified in the cryo-EM structure of NfMCU.
Extended Data Fig. 8 Western blot analysis of NfMCU expression in E. coli.
NfMCU, with its mitochondrial targeting sequence removed and replaced with a 6×His tag, was expressed in E. coli for functional analysis. Mouse anti-His antibodies (Qiagen) were used to detect NfMCU expression and mouse anti-RpoA targeting E. coli RNA polymerase alpha (Santa Cruz Biotechnology) was used for loading control. See Supplementary Fig. 1 for gel source data. The experiment was repeated three times independently with similar results.
Supplementary information
Supplementary Figure 1
This file contains the uncropped blots.
Rights and permissions
About this article
Cite this article
Nguyen, N.X., Armache, JP., Lee, C. et al. Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature 559, 570–574 (2018). https://doi.org/10.1038/s41586-018-0333-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0333-6
This article is cited by
-
Direct cell extraction of membrane proteins for structure–function analysis
Scientific Reports (2023)
-
MiR-25 overexpression inhibits titanium particle-induced osteoclast differentiation via down-regulation of mitochondrial calcium uniporter in vitro
Journal of Orthopaedic Surgery and Research (2022)
-
Discovery of EMRE in fungi resolves the true evolutionary history of the mitochondrial calcium uniporter
Nature Communications (2020)
-
Structure and mechanism of the mitochondrial Ca2+ uniporter holocomplex
Nature (2020)
-
MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth
Signal Transduction and Targeted Therapy (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.