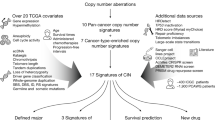
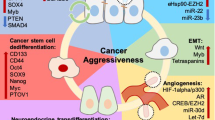
Abstract
Metastatic prostate cancer remains an incurable lethal disease. Studies indicate that prostate cancer accumulates genomic changes during disease progression and displays the highest levels of chromosomal instability (CIN) across all types of metastatic tumours. CIN, which refers to ongoing chromosomal DNA gain or loss during mitosis, and derived aneuploidy, are known to be associated with increased tumour heterogeneity, metastasis and therapy resistance in many tumour types. Paradoxically, high CIN levels are also proposed to be detrimental to tumour cell survival, suggesting that cancer cells must develop adaptive mechanisms to ensure their survival. In the context of prostate cancer, studies indicate that CIN has a key role in disease progression and might also offer a therapeutic vulnerability that can be pharmacologically targeted. Thus, a comprehensive evaluation of the causes and consequences of CIN in prostate cancer, its contribution to aggressive advanced disease and a better understanding of the acquired CIN tolerance mechanisms can translate into new tumour classifications, biomarker development and therapeutic strategies.
Key points
-
Metastatic prostate cancer displays high levels of chromosomal instability (CIN); however, the causal mechanisms and cellular and tumoural consequences of harbouring highly complex genomes are not completely understood.
-
High-CIN prostate cancer adapts to ongoing chromosomal aberration accumulation developing unique functional dependencies.
-
CIN adaptation vulnerabilities can be exploited to therapeutically target prostate cancer.
-
Evaluation of CIN levels in tumour samples and liquid biopsies might enable stratification of patient prognosis and help to personalize treatment decisions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Boutros, P. C. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 47, 736–745 (2015).
Fraser, M. et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 (2017).
Armenia, J. et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018).
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Rebello, R. J. et al. Prostate cancer. Nat. Rev. Dis. Primers 7, 9 (2021).
Hieronymus, H. et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife 7, e37294 (2018).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Viswanathan, S. R. et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 174, 433–447.e19 (2018).
Quigley, D. A. et al. Genomic hallmarks and structural variation in metastatic prostate. Cancer Cell 175, 889 (2018).
Abida, W. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Ryan, M. J. & Bose, R. Genomic alteration burden in advanced prostate cancer and therapeutic implications. Front. Oncol. 9, 1287 (2019).
Williams, J. L., Greer, P. A. & Squire, J. A. Recurrent copy number alterations in prostate cancer: an in silico meta-analysis of publicly available genomic data. Cancer Genet. 207, 474–488 (2014).
Hieronymus, H. et al. Copy number alteration burden predicts prostate cancer relapse. Proc. Natl Acad. Sci. USA 111, 11139–11144 (2014).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013).
Ben-David, U. & Amon, A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2020).
Bakhoum, S. F. & Cantley, L. C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 (2018).
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 15, 139–150 (2018).
Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018).
Alcaraz, A. et al. Aneuploidy and aneusomy of chromosome 7 detected by fluorescence in situ hybridization are markers of poor prognosis in prostate cancer. Cancer Res. 54, 3998–4002 (1994).
Stopsack, K. H. et al. Aneuploidy drives lethal progression in prostate cancer. Proc. Natl Acad. Sci. USA 116, 11390–11395 (2019).
Dhital, B. et al. Harnessing transcriptionally driven chromosomal instability adaptation to target therapy-refractory lethal prostate cancer. Cell Rep. Med. 4, 100937 (2023).
Martinez, M. J., Lyles, R. D. Z., Peinetti, N., Grunfeld, A. M. & Burnstein, K. L. Inhibition of the serine/threonine kinase BUB1 reverses taxane resistance in prostate cancer. iScience 26, 107681 (2023).
Thompson, S. L., Bakhoum, S. F. & Compton, D. A. Mechanisms of chromosomal instability. Curr. Biol. 20, R285–R295 (2010).
Bakhoum, S. F. et al. The mitotic origin of chromosomal instability. Curr. Biol. 24, R148–R149 (2014).
Gollin, S. M. Mechanisms leading to chromosomal instability. Semin. Cancer Biol. 15, 33–42 (2005).
Cortés-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 52, 331–341 (2020).
Zhang, C.-Z. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015).
Shoshani, O. et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 591, 137–141 (2021).
Ly, P. et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 51, 705–715 (2019).
Bakhoum, S. F. & Landau, D. A. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 7, a029611 (2017).
Thompson, S. L. & Compton, D. A. Chromosomes and cancer cells. Chromosome Res. 19, 433–444 (2011).
Bakhoum, S. F. & Compton, D. A. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest. 122, 1138–1143 (2012).
van Jaarsveld, R. H. & Kops, G. J. P. L. Difference makers: chromosomal instability versus aneuploidy in cancer. Trends Cancer 2, 561–571 (2016).
Tijhuis, A. E., Johnson, S. C. & McClelland, S. E. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol. Cytogenet. 12, 17 (2019).
Dhital, B. & Rodriguez-Bravo, V. Mechanisms of chromosomal instability (CIN) tolerance in aggressive tumors: surviving the genomic chaos. Chromosome Res. 31, 15 (2023).
Bakhoum, S. F., Genovese, G. & Compton, D. A. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 19, 1937–1942 (2009).
Bakhoum, S. F., Thompson, S. L., Manning, A. L. & Compton, D. A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11, 27–35 (2009).
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009).
Davoli, T., Denchi, E. L. & de Lange, T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141, 81–93 (2010).
Barber, T. D. et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl Acad. Sci. USA 105, 3443–3448 (2008).
Rodriguez-Bravo, V. et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 156, 1017–1031 (2014).
Janssen, A., van der Burg, M., Szuhai, K., Kops, G. J. P. L. & Medema, R. H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333, 1895–1898 (2011).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instabilities in human cancers. Nature 396, 643–649 (1998).
Bakhoum, S. F., Kabeche, L., Murnane, J. P., Zaki, B. I. & Compton, D. A. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 4, 1281–1289 (2014).
Nyberg, K. A., Michelson, R. J., Putnam, C. W. & Weinert, T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656 (2002).
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kaçmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004).
McAinsh, A. D. & Kops, G. J. P. L. Principles and dynamics of spindle assembly checkpoint signalling. Nat. Rev. Mol. Cell Biol. 24, 543–559 (2023).
Lara-Gonzalez, P., Westhorpe, F. G. & Taylor, S. S. The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012).
Musacchio, A. & Salmon, E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 (2007).
Bharadwaj, R. & Yu, H. The spindle checkpoint, aneuploidy, and cancer. Oncogene 23, 2016–2027 (2004).
Kops, G. J. P. L., Weaver, B. A. A. & Cleveland, D. W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5, 773–785 (2005).
Lukas, J., Lukas, C. & Bartek, J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169 (2011).
Rouse, J. & Jackson, S. P. Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551 (2002).
Lanz, M. C., Dibitetto, D. & Smolka, M. B. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 38, e101801 (2019).
Saurin, A. T. Kinase and phosphatase cross-talk at the kinetochore. Front. Cell Dev. Biol. 6, 62 (2018).
Nilsson, J. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 218, 395–409 (2019).
Schvartzman, J.-M., Sotillo, R. & Benezra, R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer 10, 102–115 (2010).
Goehring, L., Huang, T. T. & Smith, D. J. Transcription-replication conflicts as a source of genome instability. Annu. Rev. Genet. 57, 157–179 (2023).
Hamperl, S. & Cimprich, K. A. Conflict resolution in the genome: how transcription and replication make it work. Cell 167, 1455–1467 (2016).
Kim, N. & Jinks-Robertson, S. Transcription as a source of genome instability. Nat. Rev. Genet. 13, 204–214 (2012).
Drews, R. M. et al. A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 (2022).
Ippolito, M. R. et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev. Cell 56, 2440–2454.e6 (2021).
Lee, A. J. X. et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011).
Salgueiro, L. et al. Acquisition of chromosome instability is a mechanism to evade oncogene addiction. EMBO Mol. Med. 12, e10941 (2020).
Swanton, C. et al. Chromosomal instability determines taxane response. Proc. Natl Acad. Sci. USA 106, 8671–8676 (2009).
Tamura, N. et al. Specific mechanisms of chromosomal instability indicate therapeutic sensitivities in high-grade serous ovarian carcinoma. Cancer Res. 80, 4946–4959 (2020).
Watkins, T. B. K. et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 587, 126–132 (2020).
Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 9, M57–M60 (1999).
Sotillo, R. et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11, 9–23 (2007).
Gao, C. et al. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl Acad. Sci. USA 113, 14793–14798 (2016).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instability in colorectal cancers. Nature 386, 623–627 (1997).
Weaver, B. A. A. & Cleveland, D. W. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18, 658–667 (2006).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Schwarz, R. F. et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 12, e1001789 (2015).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006).
Venkatesan, S. et al. Induction of APOBEC3 exacerbates DNA replication stress and chromosomal instability in early breast and lung cancer evolution. Cancer Discov. 11, 2456–2473 (2021).
Miller, E. T. et al. Chromosomal instability in untreated primary prostate cancer as an indicator of metastatic potential. BMC Cancer 20, 398 (2020).
Funk, L. C., Zasadil, L. M. & Weaver, B. A. Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell 39, 638–652 (2016).
Siegel, J. J. & Amon, A. New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28, 189–214 (2012).
Torres, E. M., Williams, B. R. & Amon, A. Aneuploidy: cells losing their balance. Genetics 179, 737–746 (2008).
Williams, B. R. et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008).
Pfau, S. J., Silberman, R. E., Knouse, K. A. & Amon, A. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes. Dev. 30, 1395–1408 (2016).
Matthews, H. K., Bertoli, C. & de Bruin, R. A. M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 23, 74–88 (2022).
Wengner, A. M. et al. Novel Mps1 kinase inhibitors with potent antitumor activity. Mol. Cancer Ther. 15, 583–592 (2016).
Mason, J. M. et al. Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl Acad. Sci. USA 114, 3127–3132 (2017).
Sarwar, S., Morozov, V. M., Purayil, H., Daaka, Y. & Ishov, A. M. Inhibition of Mps1 kinase enhances taxanes efficacy in castration resistant prostate cancer. Cell Death Dis. 13, 868 (2022).
Malumbres, M. & Barbacid, M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009).
Stice, J. P. et al. CDK4/6 therapeutic intervention and viable alternative to taxanes in CRPC. Mol. Cancer Res. 15, 660–669 (2017).
Gong, X. et al. Genomic aberrations that activate D-type cyclins are associated with enhanced sensitivity to the CDK4 and CDK6 inhibitor abemaciclib. Cancer Cell 32, 761–776.e6 (2017).
Weaver, B. A. A., Silk, A. D., Montagna, C., Verdier-Pinard, P. & Cleveland, D. W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007).
Silk, A. D. et al. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl Acad. Sci. USA 110, E4134–E4141 (2013).
Hoevenaar, W. H. M. et al. Degree and site of chromosomal instability define its oncogenic potential. Nat. Commun. 11, 1501 (2020).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575.e11 (2022).
Lapointe, J. et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 67, 8504–8510 (2007).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Wang, Z. et al. Significance of the TMPRSS2:ERG gene fusion in prostate cancer. Mol. Med. Rep. 16, 5450–5458 (2017).
Demichelis, F. et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene 26, 4596–4599 (2007).
Visakorpi, T. et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 55, 342–347 (1995).
Camacho, N. et al. Appraising the relevance of DNA copy number loss and gain in prostate cancer using whole genome DNA sequence data. PLoS Genet. 13, e1007001 (2017).
Lalonde, E. et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 15, 1521–1532 (2014).
Lalonde, E. et al. Translating a prognostic DNA genomic classifier into the clinic: retrospective validation in 563 localized prostate tumors. Eur. Urol. 72, 22–31 (2017).
Federer-Gsponer, J. R. et al. Delineation of human prostate cancer evolution identifies chromothripsis as a polyclonal event and FKBP4 as a potential driver of castration resistance. J. Pathol. 245, 74–84 (2018).
Wu, C. et al. Poly-gene fusion transcripts and chromothripsis in prostate cancer. Genes. Chromosomes Cancer 51, 1144–1153 (2012).
Haffner, M. C. et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 18, 79–92 (2021).
Espiritu, S. M. G. et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 173, 1003–1013.e15 (2018).
Beheshti, B., Karaskova, J., Park, P. C., Squire, J. A. & Beatty, B. G. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential Giemsa banding and spectral karyotyping. Mol. Diagn. 5, 23–32 (2000).
Pan, Y. et al. Characterization of chromosomal abnormalities in prostate cancer cell lines by spectral karyotyping. Cytogenet. Cell Genet. 87, 225–232 (1999).
Ozen, M. & Pathak, S. Genetic alterations in human prostate cancer: a review of current literature. Anticancer. Res. 20, 1905–1912 (2000).
Tomlins, S. A. et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 39, 41–51 (2007).
Yu, Y. P. et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 22, 2790–2799 (2004).
Cai, C. et al. ERG induces androgen receptor-mediated regulation of SOX9. Prostate Cancer J. Clin. Invest. 123, 1109–1122 (2013).
Pihan, G. A. et al. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 61, 2212–2219 (2001).
Wang, M., Nagle, R. B., Knudsen, B. S., Cress, A. E. & Rogers, G. C. Centrosome loss results in an unstable genome and malignant prostate tumors. Oncogene 39, 399–413 (2020).
Tarapore, P. et al. Exposure to bisphenol A correlates with early-onset prostate cancer and promotes centrosome amplification and anchorage-independent growth in vitro. PLoS ONE 9, e90332 (2014).
Chan, J. Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 7, 1122–1144 (2011).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Deng, L. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).
Liu, S., Alabi, B. R., Yin, Q. & Stoyanova, T. Molecular mechanisms underlying the development of neuroendocrine prostate cancer. Semin. Cancer Biol. 86, 57–68 (2022).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Marumoto, T., Zhang, D. & Saya, H. Aurora-A — a guardian of poles. Nat. Rev. Cancer 5, 42–50 (2005).
Vader, G. & Lens, S. M. A. The Aurora kinase family in cell division and cancer. Biochim. Biophys. Acta 1786, 60–72 (2008).
Ali, A. & Stukenberg, P. T. Aurora kinases: generators of spatial control during mitosis. Front. Cell Dev. Biol. 11, 1139367 (2023).
Willems, E. et al. The functional diversity of Aurora kinases: a comprehensive review. Cell Div. 13, 7 (2018).
van der Waal, M. S., Hengeveld, R. C. C., van der Horst, A. & Lens, S. M. A. Cell division control by the chromosomal passenger complex. Exp. Cell Res. 318, 1407–1420 (2012).
Beltran, H. et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 1, 487–495 (2011).
Flores-Morales, A. et al. Proteogenomic characterization of patient-derived xenografts highlights the role of REST in neuroendocrine differentiation of castration-resistant prostate cancer. Clin. Cancer Res. 25, 595–608 (2019).
Coleman, I. M. et al. Therapeutic implications for intrinsic phenotype classification of metastatic castration-resistant prostate cancer. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 28, 3127–3140 (2022).
Hsu, E.-C. et al. Trop2 is a driver of metastatic prostate cancer with neuroendocrine phenotype via PARP1. Proc. Natl Acad. Sci. USA 117, 2032–2042 (2020).
Corella, A. N. et al. Identification of therapeutic vulnerabilities in small-cell neuroendocrine prostate cancer. Clin. Cancer Res. 26, 1667–1677 (2020).
Strominger, R. N., McGiffen, J. E. & Strominger, N. L. Morphometric and experimental studies of the red nucleus in the albino rat. Anat. Rec. 219, 420–428 (1987).
Hsu, E.-C. et al. MCM2-7 complex is a novel druggable target for neuroendocrine prostate cancer. Sci. Rep. 11, 13305 (2021).
Rotinen, M. et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 24, 1887–1898 (2018).
Dardenne, E. et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30, 563–577 (2016).
Agustinus, A. S. et al. Epigenetic dysregulation from chromosomal transit in micronuclei. Nature 619, 176–183 (2023).
Mosquera, J. M. et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia N. Y. N. 15, 1–10 (2013).
Balanis, N. G. et al. Pan-cancer convergence to a small-cell neuroendocrine phenotype that shares susceptibilities with hematological malignancies. Cancer Cell 36, 17–34.e7 (2019).
Antonarakis, E. S., Gomella, L. G. & Petrylak, D. P. When and how to use PARP inhibitors in prostate cancer: a systematic review of the literature with an update on on-going trials. Eur. Urol. Oncol. 3, 594–611 (2020).
Karanika, S., Karantanos, T., Li, L., Corn, P. G. & Thompson, T. C. DNA damage response and prostate cancer: defects, regulation and therapeutic implications. Oncogene 34, 2815–2822 (2015).
Carceles-Cordon, M. et al. Cellular rewiring in lethal prostate cancer: the architect of drug resistance. Nat. Rev. Urol. 17, 292–307 (2020).
Jordan, M. A. & Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265 (2004).
Dominguez-Brauer, C. et al. Targeting mitosis in cancer: emerging strategies. Mol. Cell 60, 524–536 (2015).
Suski, J. M., Braun, M., Strmiska, V. & Sicinski, P. Targeting cell-cycle machinery in cancer. Cancer Cell 39, 759–778 (2021).
Chang, L., Ruiz, P., Ito, T. & Sellers, W. R. Targeting pan-essential genes in cancer: challenges and opportunities. Cancer Cell 39, 466–479 (2021).
Malumbres, M. et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118, 493–504 (2004).
Salvador-Barbero, B. et al. CDK4/6 inhibitors impair recovery from cytotoxic chemotherapy in pancreatic adenocarcinoma. Cancer Cell 38, 584 (2020).
Goel, S., Bergholz, J. S. & Zhao, J. J. Targeting CDK4 and CDK6 in cancer. Nat. Rev. Cancer 22, 356–372 (2022).
Vitale, I., Galluzzi, L., Castedo, M. & Kroemer, G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385–392 (2011).
Janssen, A., Kops, G. J. P. L. & Medema, R. H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. USA 106, 19108–19113 (2009).
Petrylak, D. P. et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N. Engl. J. Med. 351, 1513–1520 (2004).
Tannock, I. F. et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 351, 1502–1512 (2004).
de Bono, J. S. et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet 376, 1147–1154 (2010).
Sweeney, C. J. et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N. Engl. J. Med. 373, 737–746 (2015).
James, N. D. et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 387, 1163–1177 (2016).
Smith, M. R. et al. Darolutamide and survival in metastatic, hormone-sensitive prostate cancer. N. Engl. J. Med. 386, 1132–1142 (2022).
Fizazi, K. et al. Abiraterone plus prednisone added to androgen deprivation therapy and docetaxel in de novo metastatic castration-sensitive prostate cancer (PEACE-1): a multicentre, open-label, randomised, phase 3 study with a 2 × 2 factorial design. Lancet 399, 1695–1707 (2022).
Nigg, E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32 (2001).
Jackson, J. R., Patrick, D. R., Dar, M. M. & Huang, P. S. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat. Rev. Cancer 7, 107–117 (2007).
Nizialek, E. & Antonarakis, E. S. PARP inhibitors in metastatic prostate cancer: evidence to date. Cancer Manag. Res. 12, 8105–8114 (2020).
Graham, L. S. et al. Mismatch repair deficiency in metastatic prostate cancer: response to PD-1 blockade and standard therapies. PLoS ONE 15, e0233260 (2020).
Fu, J., Bian, M., Jiang, Q. & Zhang, C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 5, 1–10 (2007).
Meraldi, P., Honda, R. & Nigg, E. A. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 14, 29–36 (2004).
Borisa, A. C. & Bhatt, H. G. A comprehensive review on Aurora kinase: small molecule inhibitors and clinical trial studies. Eur. J. Med. Chem. 140, 1–19 (2017).
Melichar, B. et al. Safety and activity of alisertib, an investigational Aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol. 16, 395–405 (2015).
Lin, J. et al. A phase I/II study of the investigational drug alisertib in combination with abiraterone and prednisone for patients with metastatic castration-resistant prostate cancer progressing on abiraterone. Oncologist 21, 1296–1297e (2016).
Beltran, H. et al. A phase II trial of the Aurora kinase A inhibitor alisertib for patients with castration-resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin. Cancer Res. 25, 43–51 (2019).
Archambault, V. & Glover, D. M. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10, 265–275 (2009).
Joukov, V. & De Nicolo, A. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11, eaar4195 (2018).
Wu, J., Ivanov, A. I., Fisher, P. B. & Fu, Z. Polo-like kinase 1 induces epithelial-to-mesenchymal transition and promotes epithelial cell motility by activating CRAF/ERK signaling. eLife 5, e10734 (2016).
Gheghiani, L., Shang, S. & Fu, Z. Targeting the PLK1-FOXO1 pathway as a novel therapeutic approach for treating advanced prostate cancer. Sci. Rep. 10, 12327 (2020).
Cristóbal, I., Rojo, F., Madoz-Gúrpide, J. & García-Foncillas, J. Cross talk between Wnt/β-Catenin and CIP2A/Plk1 signaling in prostate cancer: promising therapeutic implications. Mol. Cell. Biol. 36, 1734–1739 (2016).
Takai, N., Hamanaka, R., Yoshimatsu, J. & Miyakawa, I. Polo-like kinases (Plks) and cancer. Oncogene 24, 287–291 (2005).
Deeraksa, A. et al. Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene 32, 2973–2983 (2013).
Weichert, W. et al. Polo-like kinase 1 is overexpressed in prostate cancer and linked to higher tumor grades. Prostate 60, 240–245 (2004).
Patterson, J. C. et al. Plk1 inhibitors and abiraterone synergistically disrupt mitosis and kill cancer cells of disparate origin independently of androgen receptor signaling. Cancer Res. 83, 219–238 (2023).
Lauzé, E. et al. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 14, 1655–1663 (1995).
Abrieu, A. et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106, 83–93 (2001).
Kwiatkowski, N. et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 6, 359–368 (2010).
Jemaà, M. et al. Characterization of novel MPS1 inhibitors with preclinical anticancer activity. Cell Death Differ. 20, 1532–1545 (2013).
Maciejowski, J. et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190, 89–100 (2010).
Daniel, J., Coulter, J., Woo, J.-H., Wilsbach, K. & Gabrielson, E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl Acad. Sci. USA 108, 5384–5389 (2011).
Chen, S. et al. Silencing TTK expression inhibits the proliferation and progression of prostate cancer. Exp. Cell Res. 385, 111669 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02138812 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02366949 (2020).
Bettencourt-Dias, M. et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207 (2005).
Habedanck, R., Stierhof, Y.-D., Wilkinson, C. J. & Nigg, E. A. The polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146 (2005).
Basto, R. et al. Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032–1042 (2008).
Holland, A. J., Lan, W., Niessen, S., Hoover, H. & Cleveland, D. W. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188, 191–198 (2010).
Singh, C. K. et al. PLK4 is upregulated in prostate cancer and its inhibition reduces centrosome amplification and causes senescence. Prostate 82, 957–969 (2022).
Mason, J. M. et al. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell 26, 163–176 (2014).
Wong, Y. L. et al. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015).
Ocasio, C. A. et al. A first generation inhibitor of human Greatwall kinase, enabled by structural and functional characterisation of a minimal kinase domain construct. Oncotarget 7, 71182–71197 (2016).
Sharp, A. et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest. 129, 192–208 (2019).
Scher, H. I. et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2, 1441–1449 (2016).
Marín-Aguilera, M. et al. Androgen receptor and its splicing variant 7 expression in peripheral blood mononuclear cells and in circulating tumor cells in metastatic castration-resistant prostate cancer. Cells 9, 203 (2020).
Magani, F. et al. Identification of an oncogenic network with prognostic and therapeutic value in prostate cancer. Mol. Syst. Biol. 14, e8202 (2018).
King, A. Could immunotherapy finally break through in prostate cancer? Nature 609, S42–S44 (2022).
Graham, L. S. & Schweizer, M. T. Mismatch repair deficiency and clinical implications in prostate cancer. Prostate 82, S37–S44 (2022).
Morel, K. L. et al. EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat. Cancer 2, 444–456 (2021).
Hong, C. et al. cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 (2022).
Macintyre, G. et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 50, 1262–1270 (2018).
Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature 606, 984–991 (2022).
Santos, A., Wernersson, R. & Jensen, L. J. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res. 43, D1140–D1144 (2015).
Lepage, C. C., Morden, C. R., Palmer, M. C. L., Nachtigal, M. W. & McManus, K. J. Detecting chromosome instability in cancer: approaches to resolve cell-to-cell heterogeneity. Cancers 11, 226 (2019).
Lynch, A. R. et al. A survey of CIN measures across mechanistic models. Preprint at bioRxiv https://doi.org/10.1101/2023.06.15.544840 (2023).
Bakker, B. et al. Predicting CIN rates from single-cell whole genome sequencing data using an in silico model. Preprint at bioRxiv https://doi.org/10.1101/2023.02.14.528596 (2023).
Garribba, L. et al. Short-term molecular consequences of chromosome mis-segregation for genome stability. Nat. Commun. 14, 1353 (2023).
Lynch, A. R., Arp, N. L., Zhou, A. S., Weaver, B. A. & Burkard, M. E. Quantifying chromosomal instability from intratumoral karyotype diversity using agent-based modeling and Bayesian inference. eLife 11, e69799 (2022).
Schonhoft, J. D. et al. Morphology-predicted large-scale transition number in circulating tumor cells identifies a chromosomal instability biomarker associated with poor outcome in castration-resistant prostate cancer. Cancer Res. 80, 4892–4903 (2020).
Beltran, H. et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin. Cancer Res. 22, 1510–1519 (2016).
Tsaur, I. et al. Aggressive variants of prostate cancer-are we ready to apply specific treatment right now? Cancer Treat. Rev. 75, 20–26 (2019).
Greene, S. B. et al. Chromosomal instability estimation based on next generation sequencing and single cell genome wide copy number variation analysis. PLoS ONE 11, e0165089 (2016).
Brown, L. C. et al. Circulating tumor cell chromosomal instability and neuroendocrine phenotype by immunomorphology and poor outcomes in men with MCRPC treated with abiraterone or enzalutamide. Clin. Cancer Res. 27, 4077–4088 (2021).
Beck, J., Urnovitz, H. B., Riggert, J., Clerici, M. & Schütz, E. Profile of the circulating DNA in apparently healthy individuals. Clin. Chem. 55, 730–738 (2009).
Schwarzenbach, H., Hoon, D. S. B. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Thierry, A. R. et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 38, 6159–6175 (2010).
Chan, K. C. A. et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 59, 211–224 (2013).
Leary, R. J. et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 4, 162ra154 (2012).
Mo, H. et al. Genome-wide chromosomal instability by cell-free DNA sequencing predicts survival in patients with metastatic breast cancer. Breast 53, 111–118 (2020).
Heitzer, E. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 5, 30 (2013).
Schütz, E. et al. Chromosomal instability in cell-free DNA is a serum biomarker for prostate cancer. Clin. Chem. 61, 239–248 (2015).
Lee, C. U. et al. Chromosomal instability in cell-free DNA as a prognostic biomarker of metastatic hormone-sensitive prostate cancer treated with androgen deprivation therapy. Eur. Urol. Focus. 9, 89–95 (2023).
Herberts, C. et al. Deep whole-genome ctDNA chronology of treatment-resistant prostate cancer. Nature 608, 199–208 (2022).
Persons, D. L. et al. Comparison of fluorescence in situ hybridization with flow cytometry and static image analysis in ploidy analysis of paraffin-embedded prostate adenocarcinoma. Hum. Pathol. 25, 678–683 (1994).
Fleskens, S. J. H. M. et al. Simultaneous assessment of DNA ploidy and biomarker expression in paraffin-embedded tissue sections. Histopathology 57, 14–26 (2010).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Liao, H. et al. Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin. Transl. Med. 10, e102 (2020).
Fernandez, G. et al. Development and validation of an AI-enabled digital breast cancer assay to predict early-stage breast cancer recurrence within 6 years. Breast Cancer Res. 24, 93 (2022).
Cordon-Cardo, C. et al. Improved prediction of prostate cancer recurrence through systems pathology. J. Clin. Invest. 117, 1876–1883 (2007).
Donovan, M. J. et al. Systems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J. Clin. Oncol. 26, 3923–3929 (2008).
Bulten, W. et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med. 28, 154–163 (2022).
Xu, Z. et al. Deep learning predicts chromosomal instability from histopathology images. iScience 24, 102394 (2021).
Acknowledgements
V.R.-B. is funded by NIH-NCI R01 CA237398. J.D.-D. is funded by NIH-NCI R01 CA207311. Both V.R.-B. and J.D.-D. are supported by the Mayo Clinic Foundation. The authors thank E. Gerner for his continued philanthropic support.
Author information
Authors and Affiliations
Contributions
V.R.-B., J.D.-D. and M.C.-C. researched data for the article and made substantial contributions to discussion of content. All authors wrote the article and reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Urology thanks Michael Haffner, Niall Corcoran and Robert Bristow for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Carceles-Cordon, M., Orme, J.J., Domingo-Domenech, J. et al. The yin and yang of chromosomal instability in prostate cancer. Nat Rev Urol (2024). https://doi.org/10.1038/s41585-023-00845-9
Accepted:
Published:
DOI: https://doi.org/10.1038/s41585-023-00845-9