Abstract
The accurate identification and stratified treatment of clinically significant early-stage prostate cancer have been ongoing concerns since the outcomes of large international prostate cancer screening trials were reported. The controversy surrounding clinical and cost benefits of prostate cancer screening has highlighted the lack of strategies for discriminating high-risk disease (that requires early treatment) from low-risk disease (that could be managed using watchful waiting or active surveillance). Advances in molecular subtyping and multiomics nanotechnology-based prostate cancer risk delineation can enable refinement of prostate cancer molecular taxonomy into clinically meaningful and treatable subtypes. Furthermore, the presence of intertumoural and intratumoural heterogeneity in prostate cancer warrants the development of novel nanodiagnostic technologies to identify clinically significant prostate cancer in a rapid, cost-effective and accurate manner. Circulating and urinary next-generation prostate cancer biomarkers for disease molecular subtyping and the newest complementary nanodiagnostic platforms for enhanced biomarker detection are promising tools for precision prostate cancer management. However, challenges in merging both aspects and clinical translation still need to be overcome.
Key points
-
The accurate identification and personalized treatment of high-grade, clinically significant prostate cancer have been ongoing concerns since the outcomes of large international prostate cancer screening trials were published.
-
The combination of next-generation prostate cancer biomarker discoveries and the emergence of companion nanodiagnostic technologies could lead to a new era of precision prostate cancer management.
-
In-depth profiling of prostate cancer has resulted in the discovery of next-generation biomarkers such as TMPRSS2–ETS fusion genes, PCA3 and SCHLAP1, which could improve molecular subtyping and risk stratification.
-
Evolving nanotechnologies such as novel nanomaterials and nanoparticles might benefit clinical translation of next-generation prostate cancer biomarkers by improving detection speed and sensitivity for development of point-of-care diagnostics.
-
Challenges for translating both novel biomarkers and nanotechnology platforms into the clinic still need to be overcome by bridging the gap between clinical and diagnostic disciplines.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018).
DeSantis, C. E. et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J. Clin. 66, 290–308 (2016).
Quigley, D. A. et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 174, 758–769 (2018).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). This cohort study provides multi-institutional clinical sequencing information that could effect treatment decisions for patients with metastatic CRPC.
Robinson, D. R. et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). This study details the complex patterns of metastatic spread arising from prostate tumours in ten patients.
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Fraser, M. et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 (2017).
Hong, M. K. H. et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 6, 6605 (2015).
Kumar, A. et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med. 22, 369–378 (2016).
Smith, B. A. et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc. Natl Acad. Sci. USA 112, E6544–E6552 (2015).
Carreira, S. et al. Tumor clone dynamics in lethal prostate cancer. Sci. Transl Med. 6, 254ra125 (2014).
Barbieri, C. E. et al. The mutational landscape of prostate cancer. Eur. Urol. 64, 567–576 (2013).
Taylor, R. A. et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat. Commun. 8, 13671 (2017).
Spratt, D. E., Zumsteg, Z. S., Feng, F. Y. & Tomlins, S. A. Translational and clinical implications of the genetic landscape of prostate cancer. Nat. Rev. Clin. Oncol. 13, 597–610 (2016). This is a review of molecular prostate cancer drivers and remaining clinical translation challenges.
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936 (2018).
Mijuskovic, M. et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br. J. Cancer 119, 96–104 (2018).
Wedge, D. C. et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat. Genet. 50, 682–692 (2018).
Attard, G. & Beltran, H. Prioritizing precision medicine for prostate cancer. Ann. Oncol. 26, 1041–1042 (2015).
Irshad, S. et al. A molecular signature predictive of indolent prostate cancer. Sci. Transl Med. 5, 202ra122 (2013).
Polascik, T. J., Oesterling, J. E. & Partin, A. W. Prostate specific antigen: a decade of discovery -what we have learned and where we are going. J. Urol. 162, 293–306 (1999).
Catalona, W. J. et al. Measurement of prostate-specific antigen in serum as a screening-test for prostate cancer. N. Engl. J. Med. 324, 1156–1161 (1991).
Bibbins-Domingo, K., Grossman, D. C. & Curry, S. J. The US preventive services task force 2017 draft recommendation statement on screening for prostate cancer: an invitation to review and comment. JAMA 317, 1949–1950 (2017).
Pinsky, P. F., Prorok, P. C. & Kramer, B. S. Prostate cancer screening -a perspective on the current state of the evidence. N. Engl. J. Med. 376, 1285–1289 (2017).
Schroeder, F. H. et al. Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 360, 1320–1328 (2009).
Andriole, G. L. et al. Mortality results from a randomized prostate-cancer screening trial. N. Engl. J. Med. 360, 1310–1319 (2009).
Schroder, F. H. et al. Screening and prostate cancer mortality: results of the European randomised study of screening for prostate cancer (ERSPC) at 13 years of follow-up. Lancet 384, 2027–2035 (2014).
Andriole, G. L. et al. Prostate cancer screening in the randomized prostate, lung, colorectal, and ovarian cancer screening trial: mortality results after 13 years of follow-up. J. Natl Cancer Inst. 104, 125–132 (2012).
Pinsky, P. F. et al. Extended mortality results for prostate cancer screening in the PLCO trial with median follow-up of 15 years. Cancer 123, 592–599 (2017).
Tsodikov, A. et al. Reconciling the effects of screening on prostate cancer mortality in the ERSPC and PLCO trials. Ann. Intern. Med. 167, 449–455 (2017).
Shoag, J. E., Mittal, S. & Hu, J. C. Reevaluating PSA testing rates in the PLCO trial. N. Engl. J. Med. 374, 1795–1796 (2016).
Pinsky, P. F. et al. Assessing contamination and compliance in the prostate component of the prostate, lung, colorectal, and ovarian (PLCO) cancer screening trial. Clin. Trials 7, 303–311 (2010).
Moyer, V. A. U.S. Preventive Services Task Force. Screening for prostate cancer: U. S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 157, 120–134 (2012).
Jemal, A. et al. Prostate cancer incidence and PSA testing patterns in relation to USPSTF screening recommendations. JAMA 314, 2054–2061 (2015).
Sammon, J. D. et al. Prostate-specific antigen screening after 2012 US preventive services task force recommendations. JAMA 314, 2075–2077 (2015).
Kearns, J. T. et al. PSA screening, prostate biopsy, and treatment of prostate cancer in the years surrounding the USPSTF recommendation against prostate cancer screening. Cancer 124, 2733–2739 (2018).
Weiner, A. B., Matulewicz, R. S., Eggener, S. E. & Schaeffer, E. M. Increasing incidence of metastatic prostate cancer in the United States (2004–2013). Prostate Cancer Prostatic Dis. 19, 395–397 (2016).
U.S. Preventive Services Task Force et al. Screening for prostate cancer: US Preventive Services Task Force recommendation statement. JAMA 319, 1901–1913 (2018).
Carter, H. B. et al. Early detection of prostate cancer: AUA guideline. J. Urol. 190, 419–426 (2013).
Gandaglia, G. et al. The problem is not what to do with indolent and harmless prostate cancer-the problem is how to avoid finding these cancers. Eur. Urol. 70, 547–548 (2016).
Hudis, C. A. Trastuzumab — mechanism of action and use in clinical practice. N. Engl. J. Med. 357, 39–51 (2007).
Carlsson, S. V. & Kattan, M. W. Prostate cancer: personalized risk — stratified screening or abandoning it altogether? Nat. Rev. Clin. Oncol. 13, 140–142 (2016).
Grasso, C. S. et al. Integrative molecular profiling of routine clinical prostate cancer specimens. Ann. Oncol. 26, 1110–1118 (2015).
Abeshouse, A. et al. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). This is a comprehensive molecular analysis of molecular heterogeneity among primary prostate cancers.
Tomlins, S. A. et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur. Urol. 68, 555–567 (2015).
Kaffenberger, S. D. & Barbieri, C. E. Molecular subtyping of prostate cancer. Curr. Opin. Urol. 26, 213–218 (2016).
Paulo, P. et al. Molecular subtyping of primary prostate cancer reveals specific and shared target genes of different ETS rearrangements. Neoplasia 14, 600–611 (2012).
Smith, S. C. & Tomlins, S. A. Prostate cancer subtyping biomarkers and outcome: is clarity emerging? Clin. Cancer Res. 20, 4733–4736 (2014).
Na, R. et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur. Urol. 71, 740–747 (2017).
Gronberg, H. et al. Prostate cancer screening in men aged 50–69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 16, 1667–1676 (2015). This is a biomarker-based screening model to examine individualized risk prediction of high-grade prostate cancer in the Swedish population.
Ström, P. et al. The Stockholm-3 model for prostate cancer detection: algorithm update, biomarker contribution, and reflex test potential. Eur. Urol. 74, 204–210 (2018).
Barbieri, C. E., Chinnaiyan, A. M., Lerner, S. P., Swanton, C. & Rubin, M. A. The emergence of precision urologic oncology: a collaborative review on biomarker-driven therapeutics. Eur. Urol. 71, 237–246 (2017).
Prensner, J. R., Rubin, M. A., Wei, J. T. & Chinnaiyan, A. M. Beyond PSA: the next generation of prostate cancer biomarkers. Sci. Transl Med. 4, 127rv3 (2012). This paper provides an introduction to the use of next-generation prostate cancer biomarkers.
Dijkstra, S., Mulders, P. F. A. & Schalken, J. A. Clinical use of novel urine and blood based prostate cancer biomarkers: a review. Clin. Biochem. 47, 889–896 (2014).
Wu, D. et al. Urinary biomarkers in prostate cancer detection and monitoring progression. Crit. Rev. Oncol. Hematol. 118, 15–26 (2017).
Velonas, V. M., Woo, H. H., dos Remedios, C. G. & Assinder, S. J. Current status of biomarkers for prostate cancer. Int. J. Mol. Sci. 14, 11034–11060 (2013).
Jedinak, A. et al. Novel non-invasive biomarkers that distinguish between benign prostate hyperplasia and prostate cancer. BMC Cancer 15, 259 (2015).
Cuzick, J. et al. Prevention and early detection of prostate cancer. Lancet Oncol. 15, E484–E492 (2014).
Hessels, D. & Schalken, J. A. Urinary biomarkers for prostate cancer: a review. Asian J. Androl. 15, 333–339 (2013).
Narayan, V. M., Konety, B. R. & Warlick, C. Novel biomarkers for prostate cancer: an evidence-based review for use in clinical practice. Int. J. Urol. 24, 352–360 (2017).
Ploussard, G. & de la Taille, A. Urine biomarkers in prostate cancer. Nat. Rev. Urol. 7, 101–109 (2010).
Prensner, J. R. & Chinnaiyan, A. M. Oncogenic gene fusions in epithelial carcinomas. Curr. Opin. Genet. Dev. 19, 82–91 (2009).
Kumar-Sinha, C., Kalyana-Sundaram, S. & Chinnaiyan, A. M. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 7, 129 (2015).
Kumar-Sinha, C., Tomlins, S. A. & Chinnaiyan, A. M. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer 8, 497–511 (2008).
Edwards, P. A. W. Fusion genes and chromosome translocations in the common epithelial cancers. J. Pathol. 220, 244–254 (2010).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Paoloni-Giacobino, A., Chen, H. M., Peitsch, M. C., Rossier, C. & Antonarakis, S. E. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics 44, 309–320 (1997).
Wilson, S. et al. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem. J. 388, 967–972 (2005).
Carver, B. S. et al. ETS rearrangements and prostate cancer initiation. Nature 457, E1 (2009).
Linn, D. E., Penney, K. L., Bronson, R. T., Mucci, L. A. & Li, Z. Deletion of interstitial genes between TMPRSS2 and ERG promotes prostate cancer progression. Cancer Res. 76, 1869–1881 (2016).
Clark, J. et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene 26, 2667–2673 (2007).
Clark, J. P. & Cooper, C. S. ETS gene fusions in prostate cancer. Nat. Rev. Urol. 6, 429–439 (2009).
Barbieri, C. E. & Rubin, M. A. Genomic rearrangements in prostate cancer. Curr. Opin. Urol. 25, 71–76 (2015).
Mani, R. S. et al. Inflammation-induced oxidative stress mediates gene fusion formation in prostate cancer. Cell Rep. 17, 2620–2631 (2016).
Adamo, P. & Ladomery, M. R. The oncogene ERG: a key factor in prostate cancer. Oncogene 35, 403–414 (2016).
Kron, K. J. et al. TMPRSS2-ERG fusion co-opts master transcription factors and activates NOTCH signaling in primary prostate cancer. Nat. Genet. 49, 1336–1345 (2017).
Bose, R. et al. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature 546, 671–675 (2017).
Kim, J., Wu, L., Zhao, J. C., Jin, H. J. & Yu, J. TMPRSS2-ERG gene fusions induce prostate tumorigenesis by modulating microRNA miR-200c. Oncogene 33, 5183–5192 (2014).
Wu, L. T. et al. ERG is a critical regulator of Wnt/LEF1 signaling in prostate cancer. Cancer Res. 73, 6068–6079 (2013).
Kedage, V. et al. An interaction with Ewing’s sarcoma breakpoint protein EWS defines a specific oncogenic mechanism of ETS factors rearranged in prostate cancer. Cell Rep. 17, 1289–1301 (2016).
Lucas, J. M. et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 4, 1310–1325 (2014).
Tian, T. V. et al. Identification of novel TMPRSS2: ERG mechanisms in prostate cancer metastasis: involvement of MMP9 and PLXNA2. Oncogene 33, 2204–2214 (2014).
St. John, J., Powell, K., Conley-LaComb, M. K. & Chinni, S. R. TMPRSS2-ERG fusion gene expression in prostate tumor cells and its clinical and biological significance in prostate cancer progression. J. Cancer Sci. Ther. 4, 94–101 (2012).
Hu, Y. et al. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin. Cancer Res. 14, 4719–4725 (2008).
Mosquera, J. M. et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin. Cancer Res. 14, 3380–3385 (2008).
Attard, G. et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69, 2912–2918 (2009).
Wang, X. J. et al. Development of peptidomimetic inhibitors of the ERG gene fusion product in prostate cancer. Cancer Cell 31, 532–548 (2017).
Chatterjee, P. et al. The TMPRSS2-ERG gene fusion blocks XRCC4-mediated nonhomologous end-joining repair and radiosensitizes prostate cancer cells to parp inhibition. Mol. Cancer Ther. 14, 1896–1906 (2015).
Furusato, B. et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 13, 228–237 (2010).
White, N. M., Feng, F. Y. & Maher, C. A. Recurrent rearrangements in prostate cancer: causes and therapeutic potential. Curr. Drug Targets 14, 450–459 (2013).
Hessels, D. & Schalken, J. A. Recurrent gene fusions in prostate cancer: their clinical implications and uses. Curr. Urol. Rep. 14, 214–222 (2013).
Kissick, H. T., Sanda, M. G., Dunn, L. K. & Arredouani, M. S. Development of a peptide-based vaccine targeting TMPRSS2:ERG fusion-positive prostate cancer. Cancer Immunol. Immunother. 62, 1831–1840 (2013).
Sanguedolce, F. et al. Urine TMPRSS2: ERG fusion transcript as a biomarker for prostate cancer: literature review. Clin. Genitourin. Cancer 14, 117–121 (2016).
Wang, J. H., Cai, Y., Ren, C. X. & Ittmann, M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 66, 8347–8351 (2006).
Wang, J. H. et al. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 68, 8516–8524 (2008).
Park, K. et al. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J. Clin. Oncol. 32, 206–211 (2014).
Culig, Z. TMPRSS:ERG fusion in prostate cancer: from experimental approaches to prognostic studies. Eur. Urol. 66, 861–862 (2014).
Young, A. et al. Correlation of urine TMPRSS2:ERG and PCA3 to ERG plus and total prostate cancer burden. Am. J. Clin. Pathol. 138, 685–696 (2012).
Tomlins, S. A. et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 3, 94ra72 (2011).
McKiernan, J. et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2, 882–889 (2016).
Day, J. R., Jost, M., Reynolds, M. A., Groskopf, J. & Rittenhouse, H. PCA3: From basic molecular science to the clinical lab. Cancer Lett. 301, 1–6 (2011).
Ferreira, L. B. et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer 12, 507 (2012).
Bussemakers, M. J. G. et al. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59, 5975–5979 (1999).
Groskopf, J. et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 52, 1089–1095 (2006).
Cui, Y. et al. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: a systematic review and meta-analysis. Sci. Rep. 6, 25776 (2016).
Lee, G. L., Dobi, A. & Srivastava, S. Diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 8, 123–124 (2011).
Deras, I. L. et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J. Urol. 179, 1587–1592 (2008).
Marks, L. S. et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 69, 532–535 (2007).
Tinzl, M., Marberger, M., Horvath, S. & Chypre, C. DD3PCA3 RNA analysis in urine—a new perspective for detecting prostate cancer. Eur. Urol. 46, 182–186 (2004).
Salami, S. S. et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol. Oncol. 31, 566–571 (2013).
Laxman, B. et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 68, 645–649 (2008).
Prensner, J. R. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398 (2013).
Prensner, J. R. et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 (2014).
Mehra, R. et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur. Urol. 70, 549–552 (2016).
Chua, M. L. K. et al. A prostate cancer “nimbosus’’: genomic instability and schlap1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur. Urol. 72, 665–674 (2017).
Yin, C. Q. et al. Molecular profiling of pooled circulating tumor cells from prostate cancer patients using a dual-antibody-functionalized microfluidic device. Anal. Chem. 90, 3744–3751 (2018).
Di Cristofano, A. & Pandolfi, P. P. The multiple roles of PTEN in tumor suppression. Cell 100, 387–390 (2000).
Yoshimoto, M. et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet. Cytogenet. 169, 128–137 (2006).
Murphy, S. J. et al. Integrated analysis of the genomic instability of PTEN in clinically insignificant and significant prostate cancer. Mod. Pathol. 29, 143–156 (2016).
Phin, S., Moore, M. W. & Cotter, P. D. Genomic rearrangements of PTEN in prostate cancer. Front. Oncol. 3, 00240 (2013).
Lotan, T. L. et al. PTEN loss detection in prostate cancer: comparison of PTEN immunohistochemistry and PTEN FISH in a large retrospective prostatectomy cohort. Oncotarget 8, 65566–65576 (2017).
Carnero, A., Blanco-Aparicio, C., Renner, O., Link, W. & Leal, J. F. M. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets 8, 187–198 (2008).
Krohn, A. et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol. 181, 401–412 (2012).
Yoshimoto, M. et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod. Pathol. 21, 1451–1460 (2008).
Han, B. et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod. Pathol. 22, 1083–1093 (2009).
Ahearn, T. U. et al. A prospective investigation of PTEN Loss and ERG expression in lethal prostate cancer. J. Natl. Cancer Inst. 108, djv346 (2016).
King, J. C. et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat. Genet. 41, 524–526 (2009).
Carver, B. S. et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 41, 619–624 (2009).
Punnoose, E. A. et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br. J. Cancer 113, 1225–1233 (2015).
Haile, S. & Sadar, M. D. Androgen receptor and its splice variants in prostate cancer. Cell. Mol. Life Sci. 68, 3971–3981 (2011).
Hu, R. et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 69, 16–22 (2009).
Hornberg, E. et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLOS ONE 6, e19059 (2011).
Ware, K. E., Garcia-Blanco, M. A., Armstrong, A. J. & Dehm, S. M. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr. Relat. Cancer 21, T87–T103 (2014).
Dehm, S. M. & Tindall, D. J. Alternatively spliced androgen receptor variants. Endocr. Relat. Cancer 18, R183–R196 (2011).
Sun, S. H. et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 120, 2715–2730 (2010).
Watson, P. A. et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc. Natl Acad. Sci. USA 107, 16759–16765 (2010).
Liu, L. L. et al. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33, 3140–3150 (2014).
Guo, Z. Y. et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 (2009).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Scher, H. I. et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 4, 1179–1186 (2018).
Azad, A. A. et al. Androgen receptor gene aberrations in circulating cell-free dna: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 21, 2315–2324 (2015).
Salvi, S. et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br. J. Cancer 112, 1717–1724 (2015).
Wyatt, A. W. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2, 1598–1606 (2016).
Smith, M. R. et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N. Engl. J. Med. 378, 1408–1418 (2018).
Hussain, M. et al. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 378, 2465–2474 (2018).
James, N. D. et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N. Engl. J. Med. 377, 338–351 (2017).
Fizazi, K. et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N. Engl. J. Med. 377, 352–360 (2017).
Van Neste, L. et al. The epigenetic promise for prostate cancer diagnosis. Prostate 72, 1248–1261 (2012).
Xu, K. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science 338, 1465–1469 (2012).
Tomlins, S. A. et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 13, 519–528 (2008).
Rubin, M. A. et al. α-Methylacyl coenzyme a racemase as a tissue biomarker for prostate cancer. JAMA 287, 1662–1670 (2002).
Meany, D. L., Zhang, Z., Sokoll, L. J., Zhang, H. & Chan, D. W. Glycoproteomics for prostate cancer detection: changes in serum PSA glycosylation patterns. J. Proteome Res. 8, 613–619 (2009).
Sartori, D. A. & Chan, D. W. Biomarkers in prostate cancer: what’s new? Curr. Opin. Oncol. 26, 259–264 (2014).
Feng, F., Schaich, M. & Hughes, L. Current and future applications of genetic prostate cancer screening in the urologic clinic. Urol. Nurs. 34, 281–289 (2014).
Catalona, W. J. et al. A multicenter study of -2 pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J. Urol. 185, 1650–1655 (2011).
Loeb, S. et al. The prostate health index selectively identifies clinically significant prostate cancer. J. Urol. 193, 1163–1169 (2015).
de la Calle, C. et al. Multicenter evaluation of the prostate health index to detect aggressive prostate cancer in biopsy naive men. J. Urol. 194, 65–72 (2015).
Vickers, A. J. et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European randomized study of prostate cancer screening in Göteborg, Sweden. BMC Med. 6, 19 (2008).
Parekh, D. J. et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur. Urol. 68, 464–470 (2015).
Braun, K., Sjoberg, D. D., Vickers, A. J., Lilja, H. & Bjartell, A. S. A four-kallikrein panel predicts high-grade cancer on biopsy: independent validation in a community cohort. Eur. Urol. 69, 505–511 (2016).
Vickers, A. et al. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J. Clin. Oncol. 28, 2493–2498 (2010).
Henrique, R. et al. Epigenetic heterogeneity of high-grade prostatic intraepithelial neoplasia: clues for clonal progression in prostate carcinogenesis. Mol. Cancer Res. 4, 1–8 (2006).
Zhou, M., Tokumaru, Y., Sidransky, D. & Epstein, J. I. Quantitative GSTP1 methylation levels correlate with Gleason grade and tumor volume in prostate needle biopsies. J. Urol. 171, 2195–2198 (2004).
Trujillo, K. A., Jones, A. C., Griffith, J. K. & Bisoffi, M. Markers of field cancerization: proposed clinical applications in prostate biopsies. Prostate Cancer 2012, 12 (2012).
Slaughter, D. P., Southwick, H. W. & Smejkal, W. "Field cancerization" in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer 6, 963–968 (1953).
Braakhuis, B. J. M., Tabor, M. P., Kummer, J. A., Leemans, C. R. & Brakenhoff, R. H. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 63, 1727–1730 (2003).
Mehrotra, J. et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate 68, 152–160 (2008).
Djavan, B., Remzi, M., Schulman, C. C., Marberger, M. & Zlotta, A. R. Repeat prostate biopsy: who, how and when?: a review. Eur. Urol. 42, (93–103 (2002).
Partin, A. W. et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J. Urol. 192, 1081–1087 (2014).
Van Neste, L. et al. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 76, 1078–1087 (2016).
Stewart, G. D. et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J. Urol. 189, 1110–1116 (2013).
Wojno, K. J. et al. Reduced rate of repeated prostate biopsies observed in ConfirmMDx clinical utility field study. Am. Health Drug Benefits 7, 129–134 (2014).
Dall’Era, M. A. et al. Utility of the Oncotype DX® prostate cancer assay in clinical practice for treatment selection in men newly diagnosed with prostate cancer: a retrospective chart review analysis. Urol. Pract. 2, 343–348 (2015).
Cullen, J. et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low-and intermediate-risk prostate cancer. Eur. Urol. 68, 123–131 (2015).
Klein, E. A. et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 66, 550–560 (2014).
Canfield, S. E. et al. A guide for clinicians in the evaluation of emerging molecular diagnostics for newly diagnosed prostate cancer. Rev. Urol. 16, 172–180 (2014).
Cuzick, J. et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 12, 245–255 (2011).
Cuzick, J. et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br. J. Cancer 106, 1095–1099 (2012).
Whitfield, M. L. et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 (2002).
Tosoian, J. J. et al. Prognostic utility of biopsy-derived cell cycle progression score in patients with national comprehensive cancer network low-risk prostate cancer undergoing radical prostatectomy: implications for treatment guidance. BJU Int. 120, 808–814 (2017).
Marrone, M., Potosky, A. L., Penson, D. & Freedman, A. N. A 22 gene-expression assay, Decipher(R) (GenomeDx Biosciences) to predict five-year risk of metastatic prostate cancer in men treated with radical prostatectomy. PLOS Curr. https://doi.org/10.1371/currents.eogt.761b81608129ed61b0b48d42c04f92a4 (2015).
Spratt, D. E. et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J. Clin. Oncol. 35, 1991–1998 (2017).
Karnes, R. J. et al. Validation of a genomic risk classifier to predict prostate cancer-specific mortality in men with adverse pathologic features. Eur. Urol. 73, 168–175 (2018).
Klein, E. A. et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology 90, 148–152 (2016).
Klein, E. A. et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur. Urol. 67, 778–786 (2015).
Badani, K. K. et al. Effect of a genomic classifier test on clinical practice decisions for patients with high-risk prostate cancer after surgery. BJU Int. 115, 419–429 (2015).
Badani, K. et al. Impact of a genomic classifier of metastatic risk on postoperative treatment recommendations for prostate cancer patients: a report from the DECIDE study group. Oncotarget 4, 600–609 (2013).
Haese, A. et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur. Urol. 54, 1081–1088 (2008).
Chun, F. K. et al. Prostate cancer gene 3 (PCA3): development and internal validation of a novel biopsy nomogram. Eur. Urol. 56, 659–667 (2009).
Ploussard, G. et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. Eur. Urol. 59, 422–429 (2011).
Roobol, M. J. et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur. Urol. 58, 475–481 (2010).
Gittelman, M. C. et al. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: a prospective multicenter clinical study. J. Urol. 190, 64–69 (2013).
Wei, J. T. et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J. Clin. Oncol. 32, 4066–4072 (2014).
Hessels, D. & Schalken, J. A. The use of PCA3 in the diagnosis of prostate cancer. Nat. Rev. Urol. 6, 255–261 (2009).
Tomlins, S. A. et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur. Urol. 70, 45–53 (2016).
Merdan, S. et al. Assessment of long-term outcomes associated with urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion at repeat biopsy. Cancer 121, 4071–4079 (2015).
Sanda, M. G. et al. Association between combined TMPRSS2:ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA Oncol. 3, 1083–1093 (2017).
Leyten, G. et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur. Urol. 65, 534–542 (2014).
Leyten, G. et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin. Cancer Res. 21, 3061–3070 (2015).
Van Neste, L. et al. Detection of high-grade prostate cancer using a urinary molecular biomarker-based risk score. Eur. Urol. 70, 740–748 (2016).
Duijvesz, D., Luider, T., Bangma, C. H. & Jenster, G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 59, 823–831 (2011).
Nilsson, J. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br. J. Cancer 100, 1603–1607 (2009).
Overbye, A. et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 6, 30357–30376 (2015).
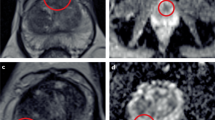
Futterer, J. J. et al. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? a systematic review of the literature. Eur. Urol. 68, 1045–1053 (2015).
Mehralivand, S. et al. A magnetic resonance imaging-based prediction model for prostate biopsy risk stratification. JAMA Oncol. 4, 678–685 (2018).
Ahmed, H. U. et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 389, 815–822 (2017).
Pokorny, M. R. et al. Prospective study of diagnostic accuracy comparing prostate cancer detection by transrectal ultrasound-guided biopsy versus magnetic resonance (MR) imaging with subsequent MR-guided biopsy in men without previous prostate biopsies. Eur. Urol. 66, 22–29 (2014).
Moore, C. M. et al. Image-guided prostate biopsy using magnetic resonance imaging-derived targets: a systematic review. Eur. Urol. 63, 125–140 (2013).
Siddiqui, M. M. et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate Cancer. JAMA 313, 390–397 (2015).
Kasivisvanathan, V. et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N. Engl. J. Med. 378, 1767–1777 (2018).
Ferrari, M. Cancer nanotechnology: opportunities and challenges. Nat. Rev. Cancer 5, 161–171 (2005).
Nie, S. M., Xing, Y., Kim, G. J. & Simons, J. W. Nanotechnology applications in cancer. Annu. Rev. Biomed. Eng. 9, 257–288 (2007). This paper gives a broad introductory review of various nanotechnologies for cancer applications.
Smith, S. J., Nemr, C. R. & Kelley, S. O. Chemistry-driven approaches for ultrasensitive nucleic acid detection. J. Am. Chem. Soc. 139, 1020–1028 (2017).
Barenholz, Y. Doxil® — the first FDA-approved nano-drug: lessons learned. J. Control. Release 160, 117–134 (2012).
Kang, B. J. et al. Diagnosis of prostate cancer via nanotechnological approach. Int. J. Nanomed. 10, 6555–6569 (2015).
Wu, G. H. et al. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat. Biotechnol. 19, 856–860 (2001). This paper details the pioneering use of nanotechnology to enhance PSA biomarker detection performance.
Zheng, G. F., Patolsky, F., Cui, Y., Wang, W. U. & Lieber, C. M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 23, 1294–1301 (2005).
Yu, X. et al. Carbon nanotube amplification strategies for highly sensitive immunodetection of cancer biomarkers. J. Am. Chem. Soc. 128, 11199–11205 (2006).
Xu, S. J., Liu, Y., Wang, T. H. & Li, J. H. Positive potential operation of a cathodic electrogenerated chemiluminescence immunosensor based on luminol and graphene for cancer biomarker detection. Anal. Chem. 83, 3817–3823 (2011).
Tilki, D., Kim, S. I., Hu, B., Dall’Era, M. A. & Evans, C. P. Ultrasensitive prostate specific antigen and its role after radical prostatectomy: a systematic review. J. Urol. 193, 1525–1531 (2015).
Grubisha, D. S., Lipert, R. J., Park, H. Y., Driskell, J. & Porter, M. D. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal. Chem. 75, 5936–5943 (2003).
Mani, V., Chikkaveeraiah, B. V., Patel, V., Gutkind, J. S. & Rusling, J. F. Ultrasensitive immunosensor for cancer biomarker proteins using gold nanoparticle film electrodes and multienzyme-particle amplification. ACS Nano 3, 585–594 (2009).
Liu, X. et al. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J. Am. Chem. Soc. 130, 2780–2782 (2008).
Wang, J., Koo, K. M., Wang, Y. & Trau, M. “Mix-to-go” silver colloidal strategy for prostate cancer molecular profiling and risk prediction. Anal. Chem. 90, 12698–12705 (2018).
Gupta, A. K., Naregalkar, R. R., Vaidya, V. D. & Gupta, M. Recent advances on surface engineering of magnetic iron oxide nanoparticles and their biomedical applications. Nanomedicine (Lond.) 2, 23–39 (2007).
Sarkar, P., Ghosh, D., Bhattacharyay, D., Setford, S. J. & Turner, A. P. F. Electrochemical immunoassay for free prostate specific antigen (f-PSA) using magnetic beads. Electroanalysis 20, 1414–1420 (2008).
Song, E. Q. et al. Fluorescent-magnetic-biotargeting multifunctional nanobioprobes for detecting and isolating multiple types of tumor cells. ACS Nano 5, 761–770 (2011).
Koo, K. M., Carrascosa, L. G., Shiddiky, M. J. A. & Trau, M. Poly(A) extensions of miRNAs for amplification-free electrochemical detection on screen-printed gold electrodes. Anal. Chem. 88, 2000–2005 (2016).
Koo, K. M., Carrascosa, L. G., Shiddiky, M. J. A. & Trau, M. Amplification-free detection of gene fusions in prostate cancer urinary samples using mRNA-gold affinity interactions. Anal. Chem. 88, 6781–6788 (2016).
Koo, K. M., Carrascosa, L. G. & Trau, M. DNA-directed assembly of copper nanoblocks with inbuilt fluorescent and electrochemical properties: application in simultaneous amplification-free analysis of multiple RNA species. Nano Res. 11, 940–952 (2018).
Koo, K. M., Wee, E. J. H. & Trau, M. Colorimetric TMPRSS2-ERG gene fusion detection in prostate cancer urinary samples via recombinase polymerase amplification. Theranostics 6, 1415–1424 (2016).
Koo, K. M., Dey, S. & Trau, M. Amplification-free multi-RNA-type profiling for cancer risk stratification via alternating current electrohydrodynamic nanomixing. Small 14, 1704025 (2018). This paper describes a nanodevice for amplification-free detection of multiple types of prostate cancer RNA biomarkers.
Koo, K. M., Wee, E. J. H., Mainwaring, P. N. & Trau, M. A simple, rapid, low-cost technique for naked-eye detection of urine-isolated TMPRSS2: ERG gene fusion RNA. Sci. Rep. 6, 30722 (2016). This paper describes a miniaturized solution-based technique for visual detection of urinary TMPRSS2–ERG status.
Koo, K. M., Dey, S. & Trau, M. A sample-to-targeted gene analysis biochip for nanofluidic manipulation of solid-phase circulating tumor nucleic acid amplification in liquid biopsies. ACS Sens. 3, 2597–2603 (2018). This paper details an integrated biochip platform for complex liquid biopsy sample-to-targeted genetic analysis.
Brus, L. Quantum crystallites and nonlinear optics. Appl. Phys. A 53, 465–474 (1991).
Chinen, A. B. et al. Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chem. Rev. 115, 10530–10574 (2015).
Li, X. et al. Rapid and quantitative detection of prostate specific antigen with a quantum dot nanobeads-based immunochromatography test strip. ACS Appl. Mater. Interfaces 6, 6406–6414 (2014).
Zhang, W., Hubbard, A., Brunhoeber, P., Wang, Y. & Tang, L. Automated multiplexing quantum dots in situ hybridization assay for simultaneous detection of ERG and PTEN gene status in prostate cancer. J. Mol. Diagn. 15, 754–764 (2013).
Fleischmann, M., Hendra, P. J. & McQuillan, A. J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 26, 163–166 (1974).
Koo, K. M., Wee, E. J. H., Wang, Y. & Trau, M. Enabling miniaturised personalised diagnostics: from lab-on-a-chip to lab-in-a-drop. Lab. Chip 17, 3200–3220 (2017). This paper provides an introduction to the concept of miniaturized ‘lab-in-a-drop’ systems.
Zong, C. et al. Surface-enhanced Raman spectroscopy for bioanalysis: reliability and challenges. Chem. Rev. 118, 4946–4980 (2018).
Cheng, Z. et al. Simultaneous detection of dual prostate specific antigens using surface-enhanced raman scattering-based immunoassay for accurate diagnosis of prostate cancer. ACS Nano 11, 4926–4933 (2017). This paper details the duplexed evaluation of free to total PSA ratio in clinical serum samples.
Koo, K. M., McNamara, B., Wee, E. J. H., Wang, Y. & Trau, M. Rapid and sensitive fusion gene detection in prostate cancer urinary specimens by label-free surface-enhanced Raman scattering. J. Biomed. Nanotechnol. 12, 1798–1805 (2016).
Koo, K. M., Wee, E. J. H., Mainwaring, P. N., Wang, Y. & Trau, M. Toward precision medicine: a cancer molecular subtyping nano-strategy for RNA biomarkers in tumor and urine. Small 12, 6233–6242 (2016). This paper details the pentaplexed detection of next-generation prostate cancer RNA biomarkers in clinical patient samples.
Wang, J., Koo, K. M., Wee, E. J. H., Wang, Y. L. & Trau, M. A nanoplasmonic label-free surface-enhanced Raman scattering strategy for non-invasive cancer genetic subtyping in patient samples. Nanoscale 9, 3496–3503 (2017).
Mir-Simon, B., Reche-Perez, I., Guerrini, L., Pazos-Perez, N. & Alvarez-Puebla, R. A. Universal one-pot and scalable synthesis of SERS encoded nanoparticles. Chem. Mater. 27, 950–958 (2015).
Kim, H. M. et al. Large scale synthesis of surface-enhanced Raman scattering nanoprobes with high reproducibility and long-term stability. J. Ind. Eng. Chem. 33, 22–27 (2016).
Whitesides, G. M. The origins and the future of microfluidics. Nature 442, 368 (2006).
Chin, C. D., Linder, V. & Sia, S. K. Commercialization of microfluidic point-of-care diagnostic devices. Lab. Chip 12, 2118–2134 (2012).
Armenia, J. et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018).
Brooks, J. D. Translational genomics: the challenge of developing cancer biomarkers. Genome Res. 22, 183–187 (2012).
Yamoah, K. et al. Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J. Clin. Oncol. 33, 2789–2796 (2015).
Simon, N. & Simon, R. Adaptive enrichment designs for clinical trials. Biostatistics 14, 613–625 (2013).
Simon, R. & Simon, N. Inference for multimarker adaptive enrichment trials. Stat. Med. 36, 4083–4093 (2017).
Koo, K. M. et al. Design and clinical verification of surface enhanced Raman spectroscopy diagnostic technology for individual cancer risk prediction. ACS Nano 12, 8362–8371 (2018). This is the first comprehensive clinical evaluation of a prostate cancer nanodiagnostic technology.
Hessels, D. et al. Analytical validation of an mRNA-based urine test to predict the presence of high-grade prostate cancer. Transl Med. Commun. 2, 5 (2017).
Acknowledgements
K.M.K. acknowledges support from the Australian Government Research Training Program Scholarship and the University of Queensland Graduate School International Travel Award. Although not directly funding this work, K.M.K., P.N.M. and M.T. acknowledge funding from the National Breast Cancer Foundation of Australia (CG 1207), Australian Research Council (DP 140104006 and DP 160102836) and Royal Brisbane Women’s Hospital Foundation (2018 Research Project Grant). These grants have considerably contributed to the environment that stimulates this work. Prostate cancer research in the laboratory of S.A.T. is supported by the US National Institutes of Health, the US Department of Defense and the Prostate Cancer Foundation. S.A.T. is supported by the A. Alfred Taubman Medical Research Institute. The authors thank all patients for their consent, which progresses the research developments mentioned herein.
Reviewer information
Nature Reviews Urology thanks T. van der Kwast and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
K.M.K. researched data for and wrote the manuscript. All authors made substantial contributions to discussion of content and reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The University of Michigan has been issued a patent on ETS gene fusions in prostate cancer on which S.A.T. is a coinventor. The diagnostic field of use has been licensed to Hologic (which acquired Gen-Probe), which has sublicensed rights to Roche (which owns Ventana Medical Systems). S.A.T. has received travel support from, and had a sponsored research agreement with, Compendia Bioscience (which was acquired by Life Technologies, which was acquired by ThermoFisher Scientific). S.A.T. has sponsored research agreements with Astellas and GenomeDx. S.A.T. has served as a consultant for and received honoraria from Roche, Ventana Medical Systems, Almac Diagnostics, Janssen, AbbVie, Sanofi and Astellas (which acquired Medivation). S.A.T. is a cofounder of, consultant for and the Laboratory Director of Strata Oncology. P.N.M. and M.T. are co-founders of XING Technologies, which has licensed intellectual property from the University of Queensland, Australia. P.N.M. is a shareholder of XING Technologies. P.N.M. has served as a consultant, advisory board member and lecturer for and received honoraria, travel support and grant support from Astellas, Bristol-Myers Squibb, Ipsen, Janssen, Medivation, Merck, Novartis, Pfizer and Genentech (a subsidiary of Roche). K.M.K. declares no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Koo, K.M., Mainwaring, P.N., Tomlins, S.A. et al. Merging new-age biomarkers and nanodiagnostics for precision prostate cancer management. Nat Rev Urol 16, 302–317 (2019). https://doi.org/10.1038/s41585-019-0178-2
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41585-019-0178-2
This article is cited by
-
Thermophoretic glycan profiling of extracellular vesicles for triple-negative breast cancer management
Nature Communications (2024)
-
Advances of medical nanorobots for future cancer treatments
Journal of Hematology & Oncology (2023)
-
Recent Progresses in Electrochemical DNA Biosensors for MicroRNA Detection
Phenomics (2022)
-
Discovery of PTN as a serum-based biomarker of pro-metastatic prostate cancer
British Journal of Cancer (2021)
-
Proteomic discovery of non-invasive biomarkers of localized prostate cancer using mass spectrometry
Nature Reviews Urology (2021)