Abstract
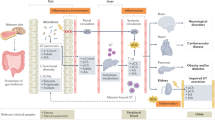
Environmental changes can induce diversity shifts within ecosystems that affect interactions between species. Similarly, the development of kidney disease induces shifts within the ecosystem of the intestinal microbiome, affecting host physiology and fitness. Renal failure itself, together with related changes in diet and medication, alters the microbiota and its secretome of micronutrients, nutrients and regulatory metabolites towards a phenotype characterized by the production of uraemic toxins, hence contributing to the clinical syndrome of uraemia and its complications. These alterations are associated with structural changes in the intestinal wall that impair barrier function and cause leakage of bacterial metabolites, bacterial wall products and live bacteria into the circulation. Thus, the intestinal microbiota represents a new therapeutic target to improve outcomes of chronic kidney disease (CKD), including symptoms of uraemia, metabolic changes, cardiovascular complications, aberrant immunity and disease progression. Initial interventional studies have shown promising effects of unselective probiotic preparations on kidney inflammation and uraemia in patients with CKD but longer-term studies are needed. Here, we take an ecological approach to understand the role of the intestinal microbiota in determining survival fitness in kidney disease.
Key points
-
The intestinal microbiota is a highly versatile ecosystem that contributes to host physiological processes, including intestinal barrier integrity, immunological fitness and metabolic fitness, and responds dynamically to intrinsic and extrinsic challenges.
-
Crosstalk between the gut microbial ecosystem and human physiological systems is context-dependent; nutrient intake and drug therapy are the most important exogenous modifiers of this crosstalk.
-
CKD can induce changes in both the composition and metabolic activity of gut microbiota (dysbiosis), with consequences for various physiological processes.
-
CKD and related changes in the microbiota impair intestinal barrier fitness and promote translocation of bacterial components into the circulation, which impairs immunological fitness by driving persistent systemic inflammation and immune paralysis.
-
CKD and related changes in the microbiota also impair metabolic and cardiovascular fitness by secreting metabolites that favour insulin resistance, obesity, endothelial dysfunction and cardiovascular ageing.
-
Faecal transplantation, specific microorganism-targeted interventions or the use of prebiotics, probiotics or dietary interventions are potential strategies to correct or manipulate CKD-related changes in the intestinal microbiota that contribute to uraemia, as well as the progression of CKD and CKD complications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kahrstrom, C. T., Pariente, N. & Weiss, U. Intestinal microbiota in health and disease. Nature 535, 47 (2016).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Thevaranjan, N. et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21, 455–466 (2017).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
Thaiss, C. A. et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551 (2016).
Vatanen, T. et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Rune, I. et al. Modulating the gut microbiota improves glucose tolerance, lipoprotein profile and atherosclerotic plaque development in ApoE-deficient mice. PLOS ONE 11, e0146439 (2016).
Skagen, K. et al. The Carnitine-butyrobetaine-trimethylamine-N-oxide pathway and its association with cardiovascular mortality in patients with carotid atherosclerosis. Atherosclerosis 247, 64–69 (2016).
Yoshida, N. et al. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 138, 2486–2498 (2018).
Zhernakova, D. V. et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat. Genet. 50, 1524–1532 (2018).
Anders, H. J., Andersen, K. & Stecher, B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 83, 1010–1016 (2013).
Andersen, K. et al. Intestinal dysbiosis, barrier dysfunction, and bacterial translocation account for CKD-related systemic inflammation. J. Am. Soc. Nephrol. 28, 76–83 (2017).
Joossens, M. et al. Gut microbiota dynamics and uraemic toxins: one size does not fit all. Gut https://doi.org/10.1136/gutjnl-2018-317561 (2018).
Poesen, R. et al. The influence of CKD on colonic microbial metabolism. J. Am. Soc. Nephrol. 27, 1389–1399 (2016).
Vaziri, N. D. et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 83, 308–315 (2013).
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. & Relman, D. A. The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262 (2012).
Dethlefsen, L., McFall-Ngai, M. & Relman, D. A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449, 811–818 (2007).
Crespo-Salgado, J. et al. Intestinal microbiota in pediatric patients with end stage renal disease: a Midwest Pediatric Nephrology Consortium study. Microbiome 4, 50 (2016).
Rakoff-Nahoum, S., Foster, K. R. & Comstock, L. E. The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016).
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
Deschasaux, M. et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 24, 1526–1531 (2018).
Cotillard, A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588 (2013).
Viaene, L. et al. Heritability and clinical determinants of serum indoxyl sulfate and p-cresyl sulfate, candidate biomarkers of the human microbiome enterotype. PLOS ONE 9, e79682 (2014).
Sonnenburg, J. L. & Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64 (2016).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Zierer, J. et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 50, 790–795 (2018).
France, M. M. & Turner, J. R. The mucosal barrier at a glance. J. Cell Sci. 130, 307–314 (2017).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Meijers, B., Farre, R., Dejongh, S., Vicario, M. & Evenepoel, P. Intestinal barrier function in chronic kidney disease. Toxins 10, E298 (2018).
Pelaseyed, T. et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20 (2014).
Johansson, M. E., Larsson, J. M. & Hansson, G. C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4659–4665 (2011).
Zarepour, M. et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 81, 3672–3683 (2013).
Mao, K. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018).
Fritz, J. H. et al. Acquisition of a multifunctional IgA+plasma cell phenotype in the gut. Nature 481, 199–203 (2011).
Nakashima, K. et al. Chitin-based barrier immunity and its loss predated mucus-colonization by indigenous gut microbiota. Nat. Commun. 9, 3402 (2018).
Joossens, M. et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637 (2011).
Leon-Coria, A., Kumar, M., Moreau, F. & Chadee, K. Defining cooperative roles for colonic microbiota and Muc2 mucin in mediating innate host defense against Entamoeba histolytica. PLOS Pathog. 14, e1007466 (2018).
Martinez-Medina, M. et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63, 116–124 (2014).
Hendrickx, A. P. et al. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of Enterococcus faecium from the intestinal epithelium. mBio 6, e01346–15 (2015).
Honda, K. & Littman, D. R. The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84 (2016).
Thaiss, C. A., Zmora, N., Levy, M. & Elinav, E. The microbiome and innate immunity. Nature 535, 65–74 (2016).
Gensollen, T., Iyer, S. S., Kasper, D. L. & Blumberg, R. S. How colonization by microbiota in early life shapes the immune system. Science 352, 539–544 (2016).
Levy, O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat. Rev. Immunol. 7, 379–390 (2007).
Rosshart, S. P. et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028 (2017).
Bach, J. F. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat. Rev. Immunol. 18, 105–120 (2018).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Diehl, G. E. et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120 (2013).
Nakajima, A. et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 215, 2019–2034 (2018).
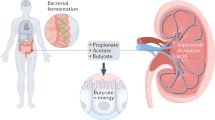
Aronov, P. A. et al. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 22, 1769–1776 (2011).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Cleophas, M. C. P. et al. Effects of oral butyrate supplementation on inflammatory potential of circulating peripheral blood mononuclear cells in healthy and obese males. Sci. Rep. 9, 775 (2019).
Marinelli, L. et al. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 9, 643 (2019).
Martin-Gallausiaux, C. et al. Butyrate produced by commensal bacteria down-regulates indolamine 2,3-dioxygenase 1 (IDO-1) expression via a dual mechanism in human intestinal epithelial cells. Front. Immunol. 9, 2838 (2018).
Masereeuw, R. et al. The kidney and uremic toxin removal: glomerulus or tubule? Semin. Nephrol. 34, 191–208 (2014).
Poesen, R. et al. Renal clearance and intestinal generation of p-cresyl sulfate and indoxyl sulfate in CKD. Clin. J. Am. Soc. Nephrol. 8, 1508–1514 (2013).
Sirich, T. L., Aronov, P. A., Plummer, N. S., Hostetter, T. H. & Meyer, T. W. Numerous protein-bound solutes are cleared by the kidney with high efficiency. Kidney Int. 84, 585–590 (2013).
Rabbers, I. et al. Metabolism at evolutionary optimal States. Metabolites 5, 311–343 (2015).
Crittenden, A. N. & Schnorr, S. L. Current views on hunter-gatherer nutrition and the evolution of the human diet. Am. J. Phys. Anthropol. 162 (Suppl. 63), 84–109 (2017).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Fragiadakis, G. K. et al. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 10, 216–227 (2018).
Schnorr, S. L. et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014).
Obregon-Tito, A. J. et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015).
Urla, C. et al. Surgical treatment of children with total colonic aganglionosis: functional and metabolic long-term outcome. BMC Surg. 18, 58 (2018).
Perry, R. J. et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534, 213–217 (2016).
Pedersen, H. K. et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016).
Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. 22, 971–982 (2015).
Baez, S. & Gordon, H. A. Tone and reactivity of vascular smooth muscle in germfree rat mesentery. J. Exp. Med. 134, 846–856 (1971).
Santisteban, M. M. et al. Hypertension-linked pathophysiological alterations in the gut. Circ. Res. 120, 312–323 (2017).
Jie, Z. et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845 (2017).
Marques, F. Z. et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 135, 964–977 (2017).
Yang, T. et al. Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–1340 (2015).
Hu, J. et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp. Mol. Med. 49, e370 (2017).
Wilck, N. et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017).
Richards, E. M., Pepine, C. J., Raizada, M. K. & Kim, S. The gut, its microbiome, and hypertension. Curr. Hypertens. Rep. 19, 36 (2017).
Jose, P. A. & Raj, D. Gut microbiota in hypertension. Curr. Opin. Nephrol. Hypertens. 24, 403–409 (2015).
Haghikia, A. et al. Gut microbiota-dependent trimethylamine N-oxide predicts risk of cardiovascular events in patients with stroke and is related to proinflammatory monocytes. Arterioscler. Thromb. Vasc. Biol. 38, 2225–2235 (2018).
Meijers, B., Jouret, F. & Evenepoel, P. Linking gut microbiota to cardiovascular disease and hypertension: lessons from chronic kidney disease. Pharmacol. Res. 133, 101–107 (2018).
Sun, C. Y., Chang, S. C. & Wu, M. S. Suppression of Klotho expression by protein-bound uremic toxins is associated with increased DNA methyltransferase expression and DNA hypermethylation. Kidney Int. 81, 640–650 (2012).
Woo, V. & Alenghat, T. Host-microbiota interactions: epigenomic regulation. Curr. Opin. Immunol. 44, 52–60 (2017).
Pluznick, J. L. Renal and cardiovascular sensory receptors and blood pressure regulation. Am. J. Physiol. Renal Physiol. 305, F439–F444 (2013).
Li, X. S. et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur. Heart J. 38, 814–824 (2017).
Tang, W. H. et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 (2015).
Tomlinson, J. A. P. & Wheeler, D. C. The role of trimethylamine N-oxide as a mediator of cardiovascular complications in chronic kidney disease. Kidney Int. 92, 809–815 (2017).
Vanholder, R., Schepers, E., Pletinck, A., Nagler, E. V. & Glorieux, G. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J. Am. Soc. Nephrol. 25, 1897–1907 (2014).
Brown, J. M. & Hazen, S. L. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu. Rev. Med. 66, 343–359 (2015).
Gregory, J. C. et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 290, 5647–5660 (2015).
Tang, W. H. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Shafi, T. et al. Trimethylamine N-oxide and cardiovascular events in hemodialysis patients. J. Am. Soc. Nephrol. 28, 321–331 (2017).
Afsar, B., Vaziri, N. D., Aslan, G., Tarim, K. & Kanbay, M. Gut hormones and gut microbiota: implications for kidney function and hypertension. J. Am. Soc. Hypertens. 10, 954–961 (2016).
Bammens, B., Verbeke, K., Vanrenterghem, Y. & Evenepoel, P. Evidence for impaired assimilation of protein in chronic renal failure. Kidney Int. 64, 2196–2203 (2003).
Evenepoel, P., Meijers, B. K., Bammens, B. R. & Verbeke, K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. Suppl. 76 (Suppl. 114), S12–S19 (2009).
Hoibian, E., Florens, N., Koppe, L., Vidal, H. & Soulage, C. O. Distal colon motor dysfunction in mice with chronic kidney disease: putative role of uremic toxins. Toxins 10, E204 (2018).
Wong, J. et al. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am. J. Nephrol. 39, 230–237 (2014).
Chiu, Y. W. et al. Pill burden, adherence, hyperphosphatemia, and quality of life in maintenance dialysis patients. Clin. J. Am. Soc. Nephrol. 4, 1089–1096 (2009).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Jiang, S. et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci. Rep. 7, 2870 (2017).
Kikuchi, M., Ueno, M., Itoh, Y., Suda, W. & Hattori, M. Uremic toxin-producing gut microbiota in rats with chronic kidney disease. Nephron 135, 51–60 (2017).
Liu, Y. et al. Disorder of gut amino acids metabolism during CKD progression is related with gut microbiota dysbiosis and metagenome change. J. Pharm. Biomed. Anal. 149, 425–435 (2018).
Nishiyama, K. et al. Chronic kidney disease after 5/6 nephrectomy disturbs the intestinal microbiota and alters intestinal motility. J. Cell. Physiol. 234, 6667–6678 (2018).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature 504, 451–455 (2013).
Mishima, E. et al. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS-based metabolomics approach. Kidney Int. 92, 634–645 (2017).
Jiang, S. et al. A reduction in the butyrate producing species Roseburia spp. and Faecalibacterium prausnitzii is associated with chronic kidney disease progression. Antonie Van Leeuwenhoek 109, 1389–1396 (2016).
Ho, W. C. & Zhang, J. Evolutionary adaptations to new environments generally reverse plastic phenotypic changes. Nat. Commun. 9, 350 (2018).
McIntyre, C. W. et al. Circulating endotoxemia: a novel factor in systemic inflammation and cardiovascular disease in chronic kidney disease. Clin. J. Am. Soc. Nephrol. 6, 133–141 (2011).
Poesen, R. et al. Associations of soluble CD14 and endotoxin with mortality, cardiovascular disease, and progression of kidney disease among patients with CKD. Clin. J. Am. Soc. Nephrol. 10, 1525–1533 (2015).
Wei, M. et al. Probiotic Bifidobacterium animalis subsp. lactis Bi-07 alleviates bacterial translocation and ameliorates microinflammation in experimental uraemia. Nephrology 19, 500–506 (2014).
Sun, L. et al. Macrophages are involved in gut bacterial translocation and reversed by Lactobacillus in experimental uremia. Dig. Dis. Sci. 61, 1534–1544 (2016).
Magnusson, M., Magnusson, K. E., Sundqvist, T. & Denneberg, T. Increased intestinal permeability to differently sized polyethylene glycols in uremic rats: effects of low- and high-protein diets. Nephron 56, 306–311 (1990).
Magnusson, M., Magnusson, K. E., Sundqvist, T. & Denneberg, T. Impaired intestinal barrier function measured by differently sized polyethylene glycols in patients with chronic renal failure. Gut 32, 754–759 (1991).
Vaziri, N. D., Yuan, J., Nazertehrani, S., Ni, Z. & Liu, S. Chronic kidney disease causes disruption of gastric and small intestinal epithelial tight junction. Am. J. Nephrol. 38, 99–103 (2013).
Vaziri, N. D., Dure-Smith, B., Miller, R. & Mirahmadi, M. K. Pathology of gastrointestinal tract in chronic hemodialysis patients: an autopsy study of 78 cases. Am. J. Gastroenterol. 80, 608–611 (1985).
Vaziri, N. D. et al. Uremic plasma impairs barrier function and depletes the tight junction protein constituents of intestinal epithelium. Am. J. Nephrol. 36, 438–443 (2012).
Vaziri, N. D., Yuan, J. & Norris, K. Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am. J. Nephrol. 37, 1–6 (2013).
Vaziri, N. D. et al. Oral activated charcoal adsorbent (AST-120) ameliorates chronic kidney disease-induced intestinal epithelial barrier disruption. Am. J. Nephrol. 37, 518–525 (2013).
Converse, R. L. Jr et al. Sympathetic overactivity in patients with chronic renal failure. N. Engl. J. Med. 327, 1912–1918 (1992).
Peschel, T. et al. Invasive assessment of bacterial endotoxin and inflammatory cytokines in patients with acute heart failure. Eur. J. Heart Fail. 5, 609–614 (2003).
Vaziri, N. D., Zhao, Y. Y. & Pahl, M. V. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol. Dial. Transplant. 31, 737–746 (2016).
Seong, E. Y., Zheng, Y., Winkelmayer, W. C., Montez-Rath, M. E. & Chang, T. I. The relationship between intradialytic hypotension and hospitalized mesenteric ischemia: a case-control study. Clin. J. Am. Soc. Nephrol. 13, 1517–1525 (2018).
Shi, K. et al. Gut bacterial translocation may aggravate microinflammation in hemodialysis patients. Dig. Dis. Sci. 59, 2109–2117 (2014).
Lau, W. L., Kalantar-Zadeh, K. & Vaziri, N. D. The gut as a source of inflammation in chronic kidney disease. Nephron 130, 92–98 (2015).
Li, L., Ma, L. & Fu, P. Gut microbiota-derived short-chain fatty acids and kidney diseases. Drug Des. Devel. Ther. 11, 3531–3542 (2017).
Pluznick, J. L. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. 90, 1191–1198 (2016).
Carlsson, A. C. et al. Soluble TNF receptors and kidney dysfunction in the elderly. J. Am. Soc. Nephrol. 25, 1313–1320 (2014).
Lai, X. et al. Outcomes of stage 1–5 chronic kidney disease in Mainland China. Ren. Fail. 36, 520–525 (2014).
Saran, R. et al. US Renal Data System 2017 annual data report: epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 71, A7 (2018).
Biswas, S. K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Hotchkiss, R. S., Coopersmith, C. M., McDunn, J. E. & Ferguson, T. A. The sepsis seesaw: tilting toward immunosuppression. Nat. Med. 15, 496–497 (2009).
Stearns-Kurosawa, D. J., Osuchowski, M. F., Valentine, C., Kurosawa, S. & Remick, D. G. The pathogenesis of sepsis. Annu. Rev. Pathol. 6, 19–48 (2011).
Einheber, A. & Carter, D. The role of the microbial flora in uremia. I. Survival times of germfree, limited-flora, and conventionalized rats after bilateral nephrectomy and fasting. J. Exp. Med. 123, 239–250 (1966).
Duranton, F. et al. Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 23, 1258–1270 (2012).
Barreto, F. C. et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin. J. Am. Soc. Nephrol. 4, 1551–1558 (2009).
Hu, J. R. et al. Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int. 94, 381–389 (2018).
Meijers, B. K. et al. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin. J. Am. Soc. Nephrol. 5, 1182–1189 (2010).
Shafi, T. et al. Serum asymmetric and symmetric dimethylarginine and morbidity and mortality in hemodialysis patients. Am. J. Kidney Dis. 70, 48–58 (2017).
Zoccali, C. et al. Plasma concentration of asymmetrical dimethylarginine and mortality in patients with end-stage renal disease: a prospective study. Lancet 358, 2113–2117 (2001).
Meijers, B. K., Bammens, B., Verbeke, K. & Evenepoel, P. A review of albumin binding in CKD. Am. J. Kidney Dis. 51, 839–850 (2008).
Klammt, S. et al. Albumin-binding capacity (ABiC) is reduced in patients with chronic kidney disease along with an accumulation of protein-bound uraemic toxins. Nephrol. Dial. Transplant. 27, 2377–2383 (2012).
Deltombe, O. et al. Exploring binding characteristics and the related competition of different protein-bound uremic toxins. Biochimie 139, 20–26 (2017).
Meijers, B. K. et al. p-Cresyl sulfate and indoxyl sulfate in hemodialysis patients. Clin. J. Am. Soc. Nephrol. 4, 1932–1938 (2009).
Kasanen, A. Serum indican and endogenous indican clearance in renal insufficiency. Ann. Med. Intern. Fenn. Suppl. 46, 1–71 (1957).
Pasternack, A., Kuhlbaeck, B. & Tallgren, L. G. Serum indican in haemodialysis. Acta. Med. Scand. 175 (Suppl. 412), 93–96 (1964).
Niwa, T. et al. Urinary indoxyl sulfate is a clinical factor that affects the progression of renal failure. Miner. Electrolyte Metab. 25, 118–122 (1999).
Niwa, T. & Ise, M. Indoxyl sulfate, a circulating uremic toxin, stimulates the progression of glomerular sclerosis. J. Lab. Clin. Med. 124, 96–104 (1994).
Niwa, T., Ise, M. & Miyazaki, T. Progression of glomerular sclerosis in experimental uremic rats by administration of indole, a precursor of indoxyl sulfate. Am. J. Nephrol. 14, 207–212 (1994).
Miyazaki, T. et al. Indoxyl sulfate stimulates renal synthesis of transforming growth factor-beta 1 and progression of renal failure. Kidney Int. Suppl. 63, S211–S214 (1997).
Dou, L. et al. The uremic solutes p-cresol and indoxyl sulfate inhibit endothelial proliferation and wound repair. Kidney Int. 65, 442–451 (2004).
Muteliefu, G., Enomoto, A., Jiang, P., Takahashi, M. & Niwa, T. Indoxyl sulphate induces oxidative stress and the expression of osteoblast-specific proteins in vascular smooth muscle cells. Nephrol. Dial. Transplant. 24, 2051–2058 (2009).
Masai, N., Tatebe, J., Yoshino, G. & Morita, T. Indoxyl sulfate stimulates monocyte chemoattractant protein-1 expression in human umbilical vein endothelial cells by inducing oxidative stress through activation of the NADPH oxidase-nuclear factor-kappaB pathway. Circ. J. 74, 2216–2224 (2010).
Fujii, H. et al. Oral charcoal adsorbent (AST-120) prevents progression of cardiac damage in chronic kidney disease through suppression of oxidative stress. Nephrol. Dial. Transplant. 24, 2089–2095 (2009).
Chiu, C. A. et al. Increased levels of total P-Cresylsulphate and indoxyl sulphate are associated with coronary artery disease in patients with diabetic nephropathy. Rev. Diabet. Stud. 7, 275–284 (2010).
Nii-Kono, T. et al. Indoxyl sulfate induces skeletal resistance to parathyroid hormone in cultured osteoblastic cells. Kidney Int. 71, 738–743 (2007).
Watanabe, K. et al. Indoxyl sulfate, a uremic toxin in chronic kidney disease, suppresses both bone formation and bone resorption. FEBS Open Bio 7, 1178–1185 (2017).
Hirata, J. et al. Indoxyl sulfate exacerbates low bone turnover induced by parathyroidectomy in young adult rats. Bone 79, 252–258 (2015).
Opdebeeck, B. et al. Indoxyl sulfate and p-Cresyl sulfate promote vascular calcification and associate with glucose intolerance. J. Am. Soc. Nephrol. 30, 751–766 (2019).
Niwa, T. et al. The protein metabolite hypothesis, a model for the progression of renal failure: an oral adsorbent lowers indoxyl sulfate levels in undialyzed uremic patients. Kidney Int. Suppl. 62, S23–S28 (1997).
Konishi, K. et al. AST-120 (Kremezin) initiated in early stage chronic kidney disease stunts the progression of renal dysfunction in type 2 diabetic subjects. Diabetes Res. Clin. Pract. 81, 310–315 (2008).
Schulman, G. et al. Randomized placebo-controlled EPPIC trials of AST-120 in CKD. J. Am. Soc. Nephrol. 26, 1732–1746 (2015).
Sato, E. et al. Impact of the oral adsorbent AST-120 on organ-specific accumulation of uremic toxins: LC-MS/MS and MS imaging techniques. Toxins 10, 19 (2018).
Koppe, L. et al. p-Cresyl sulfate promotes insulin resistance associated with CKD. J. Am. Soc. Nephrol. 24, 88–99 (2013).
Buchanan, C. et al. Intradialytic cardiac magnetic resonance imaging to assess cardiovascular responses in a short-term trial of hemodiafiltration and hemodialysis. J. Am. Soc. Nephrol. 28, 1269–1277 (2017).
Velasquez, M. T., Centron, P., Barrows, I., Dwivedi, R. & Raj, D. S. Gut microbiota and cardiovascular uremic toxicities. Toxins 10, E287 (2018).
Kolachalama, V. B. et al. Uremic solute-aryl hydrocarbon receptor-tissue factor axis associates with thrombosis after vascular injury in humans. J. Am. Soc. Nephrol. 29, 1063–1072 (2018).
Shafi, T. et al. Results of the HEMO Study suggest that p-cresol sulfate and indoxyl sulfate are not associated with cardiovascular outcomes. Kidney Int. 92, 1484–1492 (2017).
Bogiatzi, C. et al. Metabolic products of the intestinal microbiome and extremes of atherosclerosis. Atherosclerosis 273, 91–97 (2018).
Evenepoel, P., Glorieux, G. & Meijers, B. p-Cresol sulfate and indoxyl sulfate: some clouds are gathering in the uremic toxin sky. Kidney Int. 92, 1323–1324 (2017).
Vanholder, R. & Glorieux, G. The intestine and the kidneys: a bad marriage can be hazardous. Clin. Kidney J. 8, 168–179 (2015).
Noel, S. et al. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin. Pract. 127, 139–143 (2014).
Levey, A. S. & James, M. T. Acute kidney injury. Ann. Intern. Med. 167, ITC66–ITC80 (2017).
Emal, D. et al. Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 28, 1450–1461 (2017).
Andrade-Oliveira, V. et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J. Am. Soc. Nephrol. 26, 1877–1888 (2015).
Kieffer, D. A. et al. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am. J. Physiol. Renal Physiol. 310, F857–F871 (2016).
Vaziri, N. D. et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLOS ONE 9, e114881 (2014).
Li, T., Gua, C., Wu, B. & Chen, Y. Increased circulating trimethylamine N-oxide contributes to endothelial dysfunction in a rat model of chronic kidney disease. Biochem. Biophys. Res. Commun. 495, 2071–2077 (2018).
Sun, G. et al. Gut microbial metabolite TMAO contributes to renal dysfunction in a mouse model of diet-induced obesity. Biochem. Biophys. Res. Commun. 493, 964–970 (2017).
Castillo-Rodriguez, E. et al. Impact of altered intestinal microbiota on chronic kidney disease progression. Toxins 10, 300 (2018).
Koppe, L. & Fouque, D. Microbiota and prebiotics modulation of uremic toxin generation. Panminerva Med. 59, 173–187 (2017).
Mishima, E. et al. Alteration of the intestinal environment by lubiprostone is associated with amelioration of adenine-induced CKD. J. Am. Soc. Nephrol. 26, 1787–1794 (2015).
Spaulding, C. N. et al. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546, 528–532 (2017).
Zhu, W. et al. Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018).
Yacoub, R. & Wyatt, C. M. Manipulating the gut microbiome to decrease uremic toxins. Kidney Int. 91, 521–523 (2017).
Rossi, M. et al. Synbiotics easing renal failure by improving gut microbiology (SYNERGY): a randomized trial. Clin. J. Am. Soc. Nephrol. 11, 223–231 (2016).
Tayebi Khosroshahi, H. et al. Effect of high amylose resistant starch (HAM-RS2) supplementation on biomarkers of inflammation and oxidative stress in hemodialysis patients: a randomized clinical trial. Hemodial. Int. 22, 492–500 (2018).
Mishima, E. et al. Canagliflozin reduces plasma uremic toxins and alters the intestinal microbiota composition in a chronic kidney disease mouse model. Am. J. Physiol. Renal Physiol. 315, F824–F833 (2018).
Jing, W. et al. Phosphate binder, ferric citrate, attenuates anemia, renal dysfunction, oxidative stress, inflammation, and fibrosis in 5/6 nephrectomized CKD rats. J. Pharmacol. Exp. Ther. 367, 129–137 (2018).
Lau, W. L. et al. The phosphate binder ferric citrate alters the gut microbiome in rats with chronic kidney disease. J. Pharmacol. Exp. Ther. 367, 452–460 (2018).
Ramezani, A. & Raj, D. S. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 25, 657–670 (2014).
Cavalcanti Neto, M. P. et al. Gut microbiota and probiotics intervention: a potential therapeutic target for management of cardiometabolic disorders and chronic kidney disease? Pharmacol. Res. 130, 152–163 (2018).
Soleimani, A. et al. Probiotic supplementation in diabetic hemodialysis patients has beneficial metabolic effects. Kidney Int. 91, 435–442 (2017).
Kelly, J. T. et al. Healthy dietary patterns and risk of mortality and ESRD in CKD: a meta-analysis of cohort studies. Clin. J. Am. Soc. Nephrol. 12, 272–279 (2017).
Saglimbene, V. M. et al. The association of Mediterranean and DASH diets with mortality in adults on hemodialysis: the DIET-HD multinational cohort study. J. Am. Soc. Nephrol. 29, 1741–1751 (2018).
Poesen, R. et al. The influence of prebiotic arabinoxylan oligosaccharides on microbiota derived uremic retention solutes in patients with chronic kidney disease: a randomized controlled trial. PLOS ONE 11, e0153893 (2016).
So, D. et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am. J. Clin. Nutr. 107, 965–983 (2018).
Evenepoel, P., Bammens, B., Verbeke, K. & Vanrenterghem, Y. Acarbose treatment lowers generation and serum concentrations of the protein-bound solute p-cresol: a pilot study. Kidney Int. 70, 192–198 (2006).
Lee, D. M. et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc. Diabetol. 17, 62 (2018).
Devlin, A. S. et al. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe 20, 709–715 (2016).
Roberts, A. B. et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 24, 1407–1417 (2018).
Ooijevaar, R. E., Terveer, E. M., Verspaget, H. W., Kuijper, E. J. & Keller, J. J. Clinical application and potential of fecal microbiota transplantation. Annu. Rev. Med. 70, 335–351 (2019).
Mu, Q. et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 5, 73 (2017).
Laue, C. et al. Effect of a yoghurt drink containing Lactobacillus strains on bacterial vaginosis in women - a double-blind, randomised, controlled clinical pilot trial. Benef. Microbes 9, 35–50 (2018).
Derwa, Y., Gracie, D. J., Hamlin, P. J. & Ford, A. C. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment. Pharmacol. Ther. 46, 389–400 (2017).
Nadelman, P., Magno, M. B., Masterson, D., da Cruz, A. G. & Maia, L. C. Are dairy products containing probiotics beneficial for oral health? A systematic review and meta-analysis. Clin. Oral Investig. 22, 2763–2785 (2018).
Degnan, F. H. The US Food and Drug Administration and probiotics: regulatory categorization. Clin. Infect. Dis. 46, S133–S136 (2008). Suppl. 2.
Enache-Angoulvant, A. & Hennequin, C. Invasive Saccharomyces infection: a comprehensive review. Clin. Infect. Dis. 41, 1559–1568 (2005).
Rescigno, M. & Di Sabatino, A. Dendritic cells in intestinal homeostasis and disease. J. Clin. Invest. 119, 2441–2450 (2009).
Acknowledgements
H.-J.A. is supported by the Heisenberg programme of the Deutsche Forschungsgemeinschaft (AN372/24-1).
Peer review information
Nature Reviews Nephrology thanks J. Spence, N. Vaziri and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors contributed to researching data for the article, discussing the article’s content, writing the article and reviewing or editing the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Microbiome
-
Ecological community of commensal, symbiotic or pathogenic microorganisms that reside inside and outside all multicellular organisms. A microbiome includes bacteria, archaea, meiofauna, fungi and viruses.
- Archaeal microbiome
-
Archaea are single-cell microorganisms with metabolic and biochemical properties that distinguish them from bacteria. The archaeal microbiome is the community of archaea residing within a host.
- Virome
-
The collection of nucleic acids (RNA and DNA) that define the viral community inside or on multicellular organisms.
- Mycobiome
-
Fungal communities inside or on multicellular organisms.
- Meiofauna
-
Small invertebrates including unicellular protozoa and helminthic worms (for example, Entamoeba, Trichomonas or Schistosoma).
- Vertical transmission
-
Transmission from mother to child.
- 16S ribosomal RNA (rRNA) pyrosequencing
-
A method of DNA sequencing in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase, which releases pyrophosphate. This method is frequently used in microbiome genotyping.
- Intestinal congestion
-
Oedema of the intestinal wall due to volume overload and/or vascular barrier dysfunction.
- Symbiosis
-
A close and long-term biological interaction between two different biological organisms, the symbionts; the interaction can be mutualistic, commensalistic or parasitic.
- Symbionts
-
Organisms that live in a symbiotic relationship with their host, for example, Escherichia coli in the human intestinal tract.
- α-Diversity
-
Mean species diversity in a single environment (that is, within a single location).
- β-Diversity
-
The number and variability of local niches in larger environments (for example, the diversity of species between samples taken from different locations within a single host or the diversity of species between different hosts), which contribute to species diversity at a larger scale.
- Transcellular transport
-
Transport across cellular barriers through cells.
- Paracellular transport
-
Transport across cellular barriers between cells.
- Secretory IgA
-
IgA secreted by plasma cells of the gut-associated lymphoid tissue into the lumen of the intestinal tract. Secretory IgA is one of several intrinsic regulators of the intestinal microbiome.
- Chordates
-
A large group of bilateral symmetric animals sharing the anatomical structure of a stiff rod of cartilage that extends along the inside of the body. All vertebrates are chordates, but chordates also include also non-vertebrate species such as sea squirts and lancelets.
- Chitin
-
A long-chain polymer made of N-acetylglucosamine that is the primary component of cell walls in fungi, the exoskeleton of crustaceans and insects and the scales of fish.
- Dysbiosis
-
Microbial imbalance or maladaptation on or inside the body, turning a previous mutual, commensal or neutral symbiosis into a harmful of dysfunctional form of symbiosis.
- Pathobionts
-
Any potentially disease-causing organisms that, under normal circumstances, live as symbionts.
- Peyer’s patches
-
Organized lymph follicles that are part of the gut-associated lymphoid tissue. In humans, Peyer’s patches are mainly found in the small intestine.
- Metagenomic studies
-
Microbiome characterization by DNA sequencing.
- Culturomics
-
High-throughput culture-based approaches to enable extensive assessment of the microbial composition.
- Phase II metabolism
-
The metabolism of xenobiotics aims to detoxify molecules that are foreign to mammalian metabolism, for example, microbial metabolites. Phase I reactions are chemical reactions that modify the molecular structure (including oxidation, reduction and hydrolysis) and often increase the reactivity of the metabolite. Phase II reactions involve conjugation of the xenobiotic to sulfate, glucuronide or glycine, which typically reduces the reactivity of the xenobiotic.
- Microbial–host co-metabolites
-
The transformation of microbial metabolites by phase I and II metabolism results in a wide array of derivatives. These metabolites require the combined action of both microbial and mammalian metabolism.
- Hirschsprung disease
-
A congenital disorder characterized by the absence of nerves from parts of the intestinal tract, causing intestinal symptoms and growth retardation.
- Short-chain fatty acids
-
(SCFAs). The end products of carbohydrate fermentation. A group of gut-flora-derived lipid metabolites with important regulatory roles in energy metabolism, hormone secretion, systemic inflammation, hypertension and cancer.
- Saccharolytic fermentation
-
The digestion of carbohydrates under anaerobic conditions, resulting in short-chain fatty acids.
- Proteolytic fermentation
-
The digestion of peptides and amino acids under anaerobic conditions.
- Pentraxins
-
A conserved family of protein complexes made of five identical proteins. Pentraxins are acute phase proteins secreted from tissue cells or cells of the innate immune system that bind to and opsonize foreign particles or dead cell structures conceptually similar to antibodies.
- Immune paralysis
-
Acquired immunodeficiency due to the long-lasting deactivation of immune cells, which can occur following a short-lasting phase of activation, for example, as described for ‘endotoxin tolerance’ that occurs upon rechallenge with bacterial endotoxin.
- Type II errors
-
An error by which an analytical test gives a negative result although the effect is indeed present (false negative).
- Myocardial stunning
-
Persistent dysfunction of the cardiac muscle upon a transient episode of ischaemia.
- Probiotics
-
Live microorganisms that are intended to provide health benefits when consumed, typically by restoring the intestinal microflora.
- Prebiotics
-
Products that are intended to restore the intestinal microflora through ingestion of nutrients that endorse the growth or activity of beneficial microorganisms.
Rights and permissions
About this article
Cite this article
Meijers, B., Evenepoel, P. & Anders, HJ. Intestinal microbiome and fitness in kidney disease. Nat Rev Nephrol 15, 531–545 (2019). https://doi.org/10.1038/s41581-019-0172-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41581-019-0172-1
This article is cited by
-
Alteration of Plasma Indoles in Polycystic Ovary Syndrome
Reproductive Sciences (2024)
-
Nanoselenium attenuates renal ischemia-reperfusion injury in rats
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Roxadustat alleviates the inflammatory status in patients receiving maintenance hemodialysis with erythropoiesis-stimulating agent resistance by increasing the short-chain fatty acids producing gut bacteria
European Journal of Medical Research (2023)
-
Washed microbiota transplantation improves renal function in patients with renal dysfunction: a retrospective cohort study
Journal of Translational Medicine (2023)
-
Gut microbiome studies in CKD: opportunities, pitfalls and therapeutic potential
Nature Reviews Nephrology (2023)