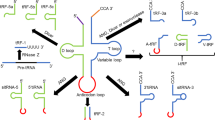
Abstract
A growing class of small RNAs, known as tRNA-derived RNAs (tdRs), tRNA-derived small RNAs or tRNA-derived fragments, have long been considered mere intermediates of tRNA degradation. These small RNAs have recently been implicated in an evolutionarily conserved repertoire of biological processes. In this Review, we discuss the biogenesis and molecular functions of tdRs in mammals, including tdR-mediated gene regulation in cell metabolism, immune responses, transgenerational inheritance, development and cancer. We also discuss the accumulation of tRNA-derived stress-induced RNAs as a distinct adaptive cellular response to pathophysiological conditions. Furthermore, we highlight new conceptual advances linking RNA modifications with tdR activities and discuss challenges in studying tdR biology in health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Schimmel, P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58 (2018).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018). This study elegantly shows a key role for pseudouridine in fine-tuning tdR-mediated regulation of protein synthesis in human embryonic and haematopoietic stem cells.
Lee, S. R. & Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 280, 42744–42749 (2005).
Thompson, D. M., Lu, C., Green, P. J. & Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 (2008). This study provides pioneering evidence that oxidative stress induces the accumulation of tdRs in different species and in human cells.
Yamasaki, S., Ivanov, P., Hu, G. F. & Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 (2009).
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014). This study shows that loss of NSUN2-mediated cytosine-5 RNA methylation leads to angiogenin-mediated cleavage of tRNAs and tdR accumulation, triggering a cellular stress response associated with neurological abnormalities in mice and humans.
Speer, J., Gehrke, C. W., Kuo, K. C., Waalkes, T. P. & Borek, E. tRNA breakdown products as markers for cancer. Cancer 44, 2120–2123 (1979).
Levitz, R. et al. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 9, 1383–1389 (1990).
Kawaji, H. et al. Hidden layers of human small RNAs. BMC Genomics 9, 157 (2008).
Aravin, A. A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 (2003).
Bühler, M., Spies, N., Bartel, D. P. & Moazed, D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15, 1015–1023 (2008).
Li, Y. et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 36, 6048–6055 (2008).
Zhang, S., Sun, L. & Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150, 378–387 (2009).
Pliatsika, V. et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 46, D152–D159 (2018).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Chen, L. et al. 5′ Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 7, eabh0494 (2021).
Kumar, P., Anaya, J., Mudunuri, S. B. & Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12, 78 (2014).
Ivanov, P., Emara, M. M., Villen, J., Gygi, S. P. & Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011). This study illustrates a role of tiRNAs in translation repression. tiRNAs inhibit translation initiation by displacing eIF4G and eIF4A from uncapped and capped RNAs.
Kim, H. K. et al. A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell Rep. 29, 3816–3824.e4 (2019). The authors unravel a crucial function of tdRs in directing mRNA translation through changes in the secondary structure of distinct ribosomal protein-encoding mRNAs.
Kim, H. K. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 (2017).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016).
Zhang, X. et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol. 196, 1591–1603 (2016).
Hanada, T. et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480 (2013).
Inoue, M. et al. Tyrosine pre-transfer RNA fragments are linked to p53-dependent neuronal cell death via PKM2. Biochem. Biophys. Res. Commun. 525, 726–732 (2020).
Cosentino, C. et al. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 46, 10302–10318 (2018).
Yeung, M. L. et al. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37, 6575–6586 (2009).
Deng, J. et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 23, 1622–1629 (2015).
Goodarzi, H. et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 (2015). The authors show that tdRs mediate a unique post-transcriptional gene regulation programme through YBX1 binding. Oncogenic response to hypoxia drives tdR accumulation, which may provide function as a tumour-suppressive mechanism in breast cancer.
Falconi, M. et al. A novel 3′-tRNA(Glu)-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J. 33, 13228–13240 (2019).
Ma, X., Liu, C. & Cao, X. Plant transfer RNA-derived fragments: biogenesis and functions. J. Integr. Plant Biol. 63, 1399–1409 (2021).
Chery, M. & Drouard, L. Plant tRNA functions beyond their major role in translation. J. Exp. Bot. 74, 2352–2363 (2023).
Shikha, S., Brogli, R., Schneider, A. & Polacek, N. tRNA biology in Trypanosomes. Chimia 73, 395–405 (2019).
Babski, J. et al. Small regulatory RNAs in Archaea. RNA Biol. 11, 484–493 (2014).
Raina, M. & Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 5, 171 (2014).
Su, Z., Wilson, B., Kumar, P. & Dutta, A. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu. Rev. Genet. 54, 47–69 (2020).
Kumar, P., Mudunuri, S. B., Anaya, J. & Dutta, A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145 (2015).
Zheng, L. L. et al. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 44, W185–W193 (2016).
Wang, J. H. et al. tsRFun: a comprehensive platform for decoding human tsRNA expression, functions and prognostic value by high-throughput small RNA-Seq and CLIP-Seq data. Nucleic Acids Res. 50, D421–D431 (2022).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Lee, Y. S., Shibata, Y., Malhotra, A. & Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 (2009).
Haussecker, D. et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 (2010).
Telonis, A. G. et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 6, 24797–24822 (2015).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Lucas, M. C. et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01743-6 (2023).
Strydom, D. J. et al. Amino acid sequence of human tumor derived angiogenin. Biochemistry 24, 5486–5494 (1985).
Pizzo, E. et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J. Cell Sci. 126, 4308–4319 (2013).
Fu, H. et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 (2009).
Su, Z., Kuscu, C., Malik, A., Shibata, E. & Dutta, A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 294, 16930–16941 (2019).
Schaefer, M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 (2010).
Tuorto, F. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905 (2012).
Vitali, P. & Kiss, T. Cooperative 2′-O-methylation of the wobble cytidine of human elongator tRNA(Met)(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev. 33, 741–746 (2019).
Li, Z. et al. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40, 6787–6799 (2012).
Thompson, D. M. & Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185, 43–50 (2009).
Thompson, D. M. & Parker, R. Stressing out over tRNA cleavage. Cell 138, 215–219 (2009).
Anderson, P. & Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 588, 4297–4304 (2014).
Cole, C. et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160 (2009).
Maute, R. L. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. USA 110, 1404–1409 (2013).
Babiarz, J. E., Ruby, J. G., Wang, Y., Bartel, D. P. & Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 (2008).
Kazimierczyk, M. et al. Characteristics of transfer RNA-derived fragments expressed during human renal cell development: the role of Dicer in tRF biogenesis. Int. J. Mol. Sci. 23, 3644 (2022).
Di Fazio, A., Schlackow, M., Pong, S. K., Alagia, A. & Gullerova, M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 50, 1734–1752 (2022).
Hasler, D. et al. The lupus autoantigen la prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol. Cell 63, 110–124 (2016).
Oberbauer, V. & Schaefer, M. R. tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. Genes 9, 607 (2018).
Kaufmann, G. Anticodon nucleases. Trends Biochem. Sci. 25, 70–74 (2000).
Donovan, J., Rath, S., Kolet-Mandrikov, D. & Korennykh, A. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 (2017).
Nechooshtan, G., Yunusov, D., Chang, K. & Gingeras, T. R. Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 48, 8035–8049 (2020).
Tosar, J. P. et al. Dimerization confers increased stability to nucleases in 5′ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 46, 9081–9093 (2018).
Costa, B. et al. Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc. Natl Acad. Sci. USA 120, e2216330120 (2023).
Jo, U. & Pommier, Y. Structural, molecular, and functional insights into Schlafen proteins. Exp. Mol. Med. 54, 730–738 (2022).
Li, M. et al. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 25, 1047–1058 (2018).
Yang, J. Y. et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA-targeting RNase engaged in translational control. Nat. Commun. 9, 1165 (2018).
Tao, E. W., Cheng, W. Y., Li, W. L., Yu, J. & Gao, Q. Y. tiRNAs: a novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J. Cell Physiol. 235, 683–690 (2020).
Dhahbi, J. M. 5′ tRNA halves: the next generation of immune signaling molecules. Front. Immunol. 6, 74 (2015).
Chen, Q. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 (2016).
Sarker, G. et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc. Natl Acad. Sci. USA 116, 10547–10556 (2019).
Sharma, U. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 (2016). This study shows that paternal diet (protein restriction) alters tdR levels during sperm maturation and regulates the expression of retroelements, thereby affecting mouse embryonic development and pre-implantation growth.
Honda, S. et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl Acad. Sci. USA 112, E3816–E3825 (2015).
Krishna, S. et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 20, e47789 (2019).
Durdevic, Z., Mobin, M. B., Hanna, K., Lyko, F. & Schaefer, M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 4, 931–937 (2013).
Li, H. et al. A dual role of human tRNA methyltransferase hTrmt13 in regulating translation and transcription. EMBO J. 41, e108544 (2022).
Hernandez-Alias, X. et al. Single-read tRNA-seq analysis reveals coordination of tRNA modification and aminoacylation and fragmentation. Nucleic Acids Res. 51, e17 (2023).
Li, J., Zhu, W. Y., Yang, W. Q., Li, C. T. & Liu, R. J. The occurrence order and cross-talk of different tRNA modifications. Sci. China Life Sci. 64, 1423–1436 (2021).
Goll, M. G. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006).
Huang, Z. X. et al. Position 34 of tRNA is a discriminative element for m5C38 modification by human DNMT2. Nucleic Acids Res. 49, 13045–13061 (2021).
Tuorto, F. et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 34, 2350–2362 (2015).
Zhang, Y. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 20, 535–540 (2018). The authors identify sperm tdR subsets, including 5′ tRNA halves, with altered abundance and RNA modifications following paternal exposure to HFD; these tdRs have direct roles in intergenerational inheritance of metabolic disorders.
Gkatza, N. A. et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 17, e3000297 (2019).
He, C. et al. TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol. 28, 62–70 (2021).
Vilardo, E. et al. Functional characterization of the human tRNA methyltransferases TRMT10A and TRMT10B. Nucleic Acids Res. 48, 6157–6169 (2020).
Deshpande, K. L. & Katze, J. R. Characterization of cDNA encoding the human tRNA-guanine transglycosylase (TGT) catalytic subunit. Gene 265, 205–212 (2001).
Chen, Y. C., Kelly, V. P., Stachura, S. V. & Garcia, G. A. Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure. RNA 16, 958–968 (2010).
Wang, X. et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 (2018).
Su, Z. et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat. Commun. 13, 2165 (2022).
Ozanick, S., Krecic, A., Andersland, J. & Anderson, J. T. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290 (2005).
Chen, Z. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47, 2533–2545 (2019).
Rashad, S. et al. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 17, 1092–1103 (2020).
Martinez, A. et al. Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 45, 5423–5436 (2017).
Reinsborough, C. W. et al. BCDIN3D regulates tRNAHis 3′ fragment processing. PLoS Genet. 15, e1008273 (2019).
Studte, P. et al. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 69, 1266–1277 (2008).
Hu, J. F. et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat. Biotechnol. 39, 978–988 (2021).
Lyons, S. M., Gudanis, D., Coyne, S. M., Gdaniec, Z. & Ivanov, P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 8, 1127 (2017).
Guzzi, N. et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 24, 299–306 (2022).
Ivanov, P. et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl Acad. Sci. USA 111, 18201–18206 (2014).
Sobala, A. & Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 10, 553–563 (2013).
Lyons, S. M. et al. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 48, 6223–6233 (2020).
Lyons, S. M., Achorn, C., Kedersha, N. L., Anderson, P. J. & Ivanov, P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 44, 6949–6960 (2016).
Gonskikh, Y. et al. Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol. 17, 1125–1136 (2020).
Goncalves, K. A. et al. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell 166, 894–906 (2016). This work reports tdR-based regulation of cell state within the mouse bone marrow niche, illustrating modulation of tdR activity in haematopoietic stem cells by angiogenin secreted from parenchymal cells.
Keam, S. P., Sobala, A., Ten Have, S. & Hutvagner, G. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 16, 413–420 (2017).
Kyriacou, S. V. & Deutscher, M. P. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol. Cell 29, 419–427 (2008).
Ying, S. et al. tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis. Signal. Transduct. Target Ther. 8, 144 (2023).
Kfoury, Y. S. et al. tiRNA signaling via stress-regulated vesicle transfer in the hematopoietic niche. Cell Stem Cell 28, 2090–2103.e9 (2021).
Kuscu, C. et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 (2018).
Diallo, I. et al. A tRNA-derived fragment present in E. coli OMVs regulates host cell gene expression and proliferation. PLoS Pathog. 18, e1010827 (2022).
Pekarsky, Y. et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl Acad. Sci. USA 113, 5071–5076 (2016).
Balatti, V. et al. tsRNA signatures in cancer. Proc. Natl Acad. Sci. USA 114, 8071–8076 (2017).
Keam, S. P. et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 42, 8984–8995 (2014).
Ruggero, K. et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 88, 3612–3622 (2014).
Schorn, A. J., Gutbrod, M. J., LeBlanc, C. & Martienssen, R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170, 61–71.e11 (2017). This study shows the role of tdRs in limiting the mobility of transposable elements in mammalian embryonic cells, revealing a conserved mechanism that controls the expression of transposons that have escaped epigenetic repression.
Sharma, U. et al. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46, 481–494.e6 (2018).
Boskovic, A., Bing, X. Y., Kaymak, E. & Rando, O. J. Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev. 34, 118–131 (2020).
Liu, X. et al. A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of its bound metabolic mRNAs. Mol. Cell 82, 2604–2617.e8 (2022).
Saikia, M. et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell Biol. 34, 2450–2463 (2014).
Cho, H. et al. Regulation of La/SSB-dependent viral gene expression by pre-tRNA 3′ trailer-derived tRNA fragments. Nucleic Acids Res. 47, 9888–9901 (2019).
Shea, J. M. et al. Genetic and epigenetic variation, but not diet, shape the sperm methylome. Dev. Cell 35, 750–758 (2015).
Alata Jimenez, N. et al. Paternal methotrexate exposure affects sperm small RNA content and causes craniofacial defects in the offspring. Nat. Commun. 14, 1617 (2023).
Natt, D. et al. Human sperm displays rapid responses to diet. PLoS Biol. 17, e3000559 (2019).
Dhahbi, J. M. et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 14, 298 (2013).
Bayazit, M. B. et al. Small RNAs derived from tRNA fragmentation regulate the functional maturation of neonatal β cells. Cell Rep. 40, 111069 (2022).
Szabat, M. et al. Musashi expression in β-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2, e232 (2011).
Jackman, J. E., Montange, R. K., Malik, H. S. & Phizicky, E. M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9, 574–585 (2003).
Zung, A. et al. Homozygous deletion of TRMT10A as part of a contiguous gene deletion in a syndrome of failure to thrive, delayed puberty, intellectual disability and diabetes mellitus. Am. J. Med. Genet. A 167A, 3167–3173 (2015).
Igoillo-Esteve, M. et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 9, e1003888 (2013).
Delaunay, S. et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 607, 593–603 (2022).
Wang, Y. et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 12, 6314 (2021).
Orellana, E. A. et al. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 81, 3323–3338.e14 (2021).
Lin, S. et al. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e5 (2018).
Scholler, E. et al. Balancing of mitochondrial translation through METTL8-mediated m(3)C modification of mitochondrial tRNAs. Mol. Cell 81, 4810–4825.e12 (2021).
Greenway, M. J. et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 38, 411–413 (2006).
Steidinger, T. U., Standaert, D. G. & Yacoubian, T. A. A neuroprotective role for angiogenin in models of Parkinson’s disease. J. Neurochem. 116, 334–341 (2011).
Schaffer, A. E. et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663 (2014).
Karaca, E. et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650 (2014).
Prehn, J. H. M. & Jirstrom, E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration. Acta Pharmacol. Sin. 41, 442–446 (2020).
Crivello, M. et al. Pleiotropic activity of systemically delivered angiogenin in the SOD1(G93A) mouse model. Neuropharmacology 133, 503–511 (2018).
Hogg, M. C. et al. Elevation in plasma tRNA fragments precede seizures in human epilepsy. J. Clin. Invest. 129, 2946–2951 (2019).
Magee, R., Londin, E. & Rigoutsos, I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 65, 203–209 (2019).
Silberstein, L. et al. Proximity-based differential single-cell analysis of the niche to identify stem/progenitor cell regulators. Cell Stem Cell 19, 530–543 (2016).
Emara, M. M. et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285, 10959–10968 (2010).
Tsuji, T. et al. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 65, 1352–1360 (2005).
Yue, T. et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 372, eaba4220 (2021).
Chiou, N. T., Kageyama, R. & Ansel, K. M. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 25, 3356–3370.e4 (2018).
Pawar, K., Shigematsu, M., Sharbati, S. & Kirino, Y. Infection-induced 5′-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 18, e3000982 (2020).
Zhang, Y. et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 6, 172–174 (2014).
Fu, M. et al. Emerging roles of tRNA-derived fragments in cancer. Mol. Cancer 22, 30 (2023).
Yu, M. et al. tRNA-derived RNA fragments in cancer: current status and future perspectives. J. Hematol. Oncol. 13, 121 (2020).
Pekarsky, Y., Balatti, V. & Croce, C. M. tRNA-derived fragments (tRFs) in cancer. J. Cell Commun. Signal. 17, 47–54 (2023).
Balatti, V. et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 112, 2169–2174 (2015).
Huang, B. et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 77, 3194–3206 (2017).
Iliopoulos, D. et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39, 761–772 (2010).
Piepoli, A. et al. MiRNA expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One 7, e33663 (2012).
Sun, V. et al. Antitumor activity of miR-1280 in melanoma by regulation of Src. Mol. Ther. 23, 71–78 (2015).
Majid, S. et al. MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS One 7, e46743 (2012).
Wang, F. et al. A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget 6, 2709–2724 (2015).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Garcia-Vilchez, R. et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol. Cancer 22, 119 (2023).
Huh, D. et al. A stress-induced tyrosine-tRNA depletion response mediates codon-based translational repression and growth suppression. EMBO J. 40, e106696 (2021).
Cui, H. et al. A novel 3′tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer.Cell Death Dis. 13, 471 (2022).
Tao, E. W. et al. A specific tRNA half, 5′tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J. Exp. Clin. Cancer Res. 40, 67 (2021).
Pan, L. et al. Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses pancreatic ductal adenocarcinoma progression. J. Clin. Invest. 131, e148130 (2021).
Watkins, C. P., Zhang, W., Wylder, A. C., Katanski, C. D. & Pan, T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 13, 2491 (2022).
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 (2016).
Miller, K. N. et al. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184, 5506–5526 (2021).
Zhao, Y., Simon, M., Seluanov, A. & Gorbunova, V. DNA damage and repair in age-related inflammation. Nat. Rev. Immunol. 23, 75–89 (2023).
Li, X. et al. tRNA-derived small RNAs: novel regulators of cancer hallmarks and targets of clinical application. Cell Death Discov. 7, 249 (2021).
Acknowledgements
We thank all the members of the Bellodi and Liu laboratory for their helpful comments. C.B. is supported by the Swedish Research Council (Vetenskapsrådet) and the Swedish Cancer Society (Cancerfonden). C.B. is a Ragnar Söderberg Fellow in Medicine and Cancerfonden Young Investigator. S.M. is a Cancerfonden postdoctoral fellow. R.J.L. is supported by the National Key Research and Development Program of China (2021YFA1100800, 2020YFA0803400) and the National Natural Science Foundation of China (31971230,32022040).
Author information
Authors and Affiliations
Contributions
All authors discussed the content, researched data and contributed to the writing of the article. C.B., S.M. and R.J.L. reviewed and edited the original draft.
Corresponding authors
Ethics declarations
Competing interests
C.B. and S.M. are founders and members of the scientific advisory board of SACRA Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Zhangli Su, Juan Tosar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
MODOMICS: https://iimcb.genesilico.pl/modomics/
Glossary
- Acceptor stem
-
A 7–9 base pair tRNA stem with a specific CCA at the 3′ end; used to accept amino acids.
- Anticodon loop
-
A structural element in tRNAs containing a 3-nucleotide sequence complementary to the corresponding codon present in the mRNA.
- Argonaute proteins
-
A family of proteins that directly bind and mediate the function of regulatory small RNAs. The eukaryotic family is divided into Argonaute proteins and PIWI proteins.
- Cajal body
-
Nuclear membraneless organelle involved in RNA metabolism and processing.
- D-arm
-
A feature in the tertiary structure of tRNAs, consisting of the D-stem and D-loop, which are named after the presence of specific dihydrouridines (D) in the D-loop.
- D-loop
-
A structural element of the D-arm in tRNA that contains dihydrouridine residues and consists of 7–11 nucleotides enclosed by a Watson and Crick base pair.
- eIF4F
-
The eukaryotic translation initiation complex, which consists of proteins that bind the 7-methylguanosine (m7G) cap structure, thereby promoting translation initiation.
- Epididymosomes
-
Membrane-bound vesicles secreted by epididymal epithelial cells; epididymosomes are vital for male fertility.
- Multi-synthetase complex
-
Also known as the aminoacyl–tRNA synthetase complex, a large complex found in higher eukaryotes, composed of multiple aminoacyl–tRNA synthetases and scaffold proteins.
- PIWI-interacting RNAs
-
PIWI-interacting RNAs are a class of non-coding small RNAs in animals, typically 21–31 nucleotides long, which interact with PIWI proteins to regulate germline differentiation and development.
- Ribotoxins
-
Protein toxins that specifically target RNA; one of the largest categories of protein toxins found in bacteria.
- TΨC loop
-
The hairpin loop nearest the 3′ end of tRNA molecules, containing thymidine (T) and pseudouridine (Ψ) bases.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muthukumar, S., Li, CT., Liu, RJ. et al. Roles and regulation of tRNA-derived small RNAs in animals. Nat Rev Mol Cell Biol 25, 359–378 (2024). https://doi.org/10.1038/s41580-023-00690-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-023-00690-z