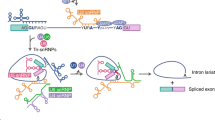
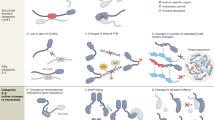
Abstract
Alternative splicing is a substantial contributor to the high complexity of transcriptomes of multicellular eukaryotes. In this Review, we discuss the accumulated evidence that most of this complexity is reflected at the protein level and fundamentally shapes the physiology and pathology of organisms. This notion is supported not only by genome-wide analyses but, mainly, by detailed studies showing that global and gene-specific modulations of alternative splicing regulate highly diverse processes such as tissue-specific and species-specific cell differentiation, thermal regulation, neuron self-avoidance, infrared sensing, the Warburg effect, maintenance of telomere length, cancer and autism spectrum disorders (ASD). We also discuss how mastering the control of alternative splicing paved the way to clinically approved therapies for hereditary diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chow, L. T., Gelinas, R. E., Broker, T. R. & Roberts, R. J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977).
Berget, S. M., Moore, C. & Sharp, P. A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA 74, 3171–3175 (1977).
Gilbert, W. Why genes in pieces? Nature 271, 501 (1978).
Khan, M. R., Wellinger, R. J. & Laurent, B. Exploring the alternative splicing of long noncoding RNAs. Trends Genet. 37, 695–698 (2021).
Rogers, S. O. Integrated evolution of ribosomal RNAs, introns, and intron nurseries. Genetica 147, 103–119 (2019).
Sekulovski, S. et al. Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia. Nat. Commun. 12, 5610 (2021).
Kearse, M. G. & Wilusz, J. E. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 31, 1717–1731 (2017).
Smith, C. W. & Valcárcel, J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388 (2000).
Kastner, B., Will, C. L., Stark, H. & Lührmann, R. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol. 11, a032417 (2019).
Licatalosi, D. D. & Darnell, R. B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11, 75–87 (2010).
Irimia, M. & Blencowe, B. J. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323–332 (2012).
Kornblihtt, A. R. et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14, 153–165 (2013).
Fu, X.-D. & Ares, M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 (2014).
Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017).
Ule, J. & Blencowe, B. J. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell 76, 329–345 (2019).
Shenasa, H. & Hertel, K. J. Combinatorial regulation of alternative splicing. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 194392 (2019).
Gordon, J. M., Phizicky, D. V. & Neugebauer, K. M. Nuclear mechanisms of gene expression control: pre-mRNA splicing as a life or death decision.Curr. Opin. Genet. Dev. 67, 67–76 (2021).
Buratti, E., Baralle, M. & Baralle, F. E. From single splicing events to thousands: the ambiguous step forward in splicing research. Brief. Funct. Genomics 12, 3–12 (2013).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Saldi, T., Riemondy, K., Erickson, B. & Bentley, D. L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell 81, 1789–1801.e5 (2021).
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R. & Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011).
de la Mata, M. et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003).
Dujardin, G. et al. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690 (2014).
Fong, N. et al. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28, 2663–2676 (2014).
Maslon, M. M. et al. A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J. 38, e101244 (2019).
Schor, I. E., Rascovan, N., Pelisch, F., Alló, M. & Kornblihtt, A. R. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl Acad. Sci. USA 106, 4325–4330 (2009).
Alló, M. et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 (2009).
Schor, I. E., Fiszbein, A., Petrillo, E. & Kornblihtt, A. R. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 32, 2264–2274 (2013).
Segelle, A. et al. Histone marks regulate the epithelial-to-mesenchymal transition via alternative splicing. Cell Rep. 38, 110357 (2022).
Eling, N., Morgan, M. D. & Marioni, J. C. Challenges in measuring and understanding biological noise. Nat. Rev. Genet. 20, 536–548 (2019).
Tress, M. L., Abascal, F. & Valencia, A. Most alternative isoforms are not functionally important. Trends Biochem. Sci. 42, 98–110 (2017).
Rodriguez, J. M., Pozo, F., di Domenico, T., Vazquez, J. & Tress, M. L. An analysis of tissue-specific alternative splicing at the protein level. PLoS Comput. Biol. 16, e1008287 (2020).
Pozo, F. et al. Assessing the functional relevance of splice isoforms. NAR Genomics Bioinform 3, lqab044 (2021).
Blencowe, B. J. The relationship between alternative splicing and proteomic complexity. Trends Biochem. Sci. 42, 407–408 (2017).
Ingolia, N. T. Ribosome footprint profiling of translation throughout the genome. Cell 165, 22–33 (2016).
Weatheritt, R. J., Sterne-Weiler, T. & Blencowe, B. J. The ribosome-engaged landscape of alternative splicing. Nat. Struct. Mol. Biol. 23, 1117–1123 (2016).
Kurosaki, T. & Maquat, L. E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 129, 461–467 (2016).
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016).
Sterne-Weiler, T. et al. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res. 23, 1615–1623 (2013).
Xing, Y. & Lee, C. Alternative splicing and RNA selection pressure — evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 7, 499–509 (2006).
Bell, L. R., Horabin, J. I., Schedl, P. & Cline, T. W. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell 65, 229–239 (1991).
Gracheva, E. O. et al. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature 476, 88–91 (2011).
Amara, S. G., Jonas, V., Rosenfeld, M. G., Ong, E. S. & Evans, R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298, 240–244 (1982).
Peterson, M. L. Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition. Wiley Interdiscip. Rev. RNA 2, 92–105 (2011).
Hattori, D. et al. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature 461, 644–648 (2009).
Emery, A. & Swanstrom, R. HIV-1: to splice or not to splice, that is the question. Viruses 13, 181–190 (2021).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
El Marabti, E. & Younis, I. The cancer spliceome: reprograming of alternative splicing in cancer. Front. Mol. Biosci. 5, 80 (2018).
Bangru, S. et al. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat. Struct. Mol. Biol. 25, 928–939 (2018).
Jensen, M. A., Wilkinson, J. E. & Krainer, A. R. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat. Struct. Mol. Biol. 21, 189–197 (2014).
Anczuków, O. & Krainer, A. R. Splicing-factor alterations in cancers. RNA 22, 1285–1301 (2016).
Cherry, S. & Lynch, K. W. Alternative splicing and cancer: insights, opportunities, and challenges from an expanding view of the transcriptome. Genes Dev. 34, 1005–1016 (2020).
Gurnari, C., Pagliuca, S. & Visconte, V. Alternative splicing in myeloid malignancies. Biomedicines 9, 1844–1851 (2021).
Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007).
Das, S., Anczuków, O., Akerman, M. & Krainer, A. R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 (2012).
Rahman, M. A., Lin, K.-T., Bradley, R. K., Abdel-Wahab, O. & Krainer, A. R. Recurrent SRSF2 mutations in MDS affect both splicing and NMD. Genes Dev. 34, 413–427 (2020).
Papasaikas, P., Tejedor, J. R., Vigevani, L. & Valcárcel, J. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell 57, 7–22 (2015).
Saltzman, A. L., Pan, Q. & Blencowe, B. J. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 25, 373–384 (2011).
Pleiss, J. A., Whitworth, G. B., Bergkessel, M. & Guthrie, C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 5, e90 (2007).
Radzisheuskaya, A. et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 26, 992–1012 (2019).
Braun, C. J. et al. Coordinated splicing of regulatory detained introns within oncogenic transcripts creates an exploitable vulnerability in malignant glioma. Cancer Cell 32, 411–426.e11 (2017).
Sanford, J. R., Gray, N. K., Beckmann, K. & Cáceres, J. F. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 (2004).
Haward, F. et al. Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. eLife 10, e65104 (2021).
Munkley, J. et al. Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer. eLife 8, e47678 (2019).
Irimia, M. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein–protein interaction networks. Mol. Cell 46, 884–892 (2012).
Buljan, M. et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell 46, 871–883 (2012).
Gonatopoulos-Pournatzis, T. et al. Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol. Cell 77, 1176–1192.e16 (2020).
Parras, A. et al. Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing. Nature 560, 441–446 (2018).
Sharon, G. et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600–1618 (2019).
Haltenhof, T. et al. A conserved kinase-based body-temperature sensor globally controls alternative splicing and gene expression. Mol. Cell 78, 57–69 (2020).
Martin Anduaga, A. et al. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. eLife 8, e44642 (2019).
Barbosa-Morais, N. L. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012).
Merkin, J., Russell, C., Chen, P. & Burge, C. B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593–1599 (2012).
Gueroussov, S. et al. An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349, 868–873 (2015).
Guo, W. et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18, 766–773 (2012).
Murphy, P. A. et al. Alternative RNA splicing in the endothelium mediated in part by Rbfox2 regulates the arterial response to low flow. eLife 7, e29494 (2018).
Vernia, S. et al. An alternative splicing program promotes adipose tissue thermogenesis. eLife 5, e17672 (2016).
Marcheva, B. et al. A role for alternative splicing in circadian control of exocytosis and glucose homeostasis. Genes Dev. 34, 1089–1105 (2020).
Ha, K. C. H., Sterne-Weiler, T., Morris, Q., Weatheritt, R. J. & Blencowe, B. J. Differential contribution of transcriptomic regulatory layers in the definition of neuronal identity. Nat. Commun. 12, 335 (2021).
Leff, S. E., Evans, R. M. & Rosenfeld, M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell 48, 517–524 (1987).
Schmucker, D. et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000).
Graveley, B. R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123, 65–73 (2005).
Goodman, K. M. et al. How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition. eLife 11, e72416 (2022).
Penev, A. et al. Alternative splicing is a developmental switch for hTERT expression. Mol. Cell 81, 2349–2360.e6 (2021).
Fiszbein, A. et al. Alternative splicing of G9a regulates neuronal differentiation. Cell Rep. 14, 2797–2808 (2016).
Gabut, M. et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146 (2011).
Han, H. et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245 (2013).
Choksi, A. et al. Tumor suppressor SMAR1 regulates PKM alternative splicing by HDAC6-mediated deacetylation of PTBP1. Cancer Metab. 9, 16–21 (2021).
David, C. J., Chen, M., Assanah, M., Canoll, P. & Manley, J. L. hnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 (2010).
Mirtschink, P. et al. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature 522, 444–449 (2015).
Li, G. et al. Exclusion of alternative exon 33 of Ca V 1.2 calcium channels in heart is proarrhythmogenic. Proc. Natl Acad. Sci. USA 114, E4288–E4295 (2017).
Sebastian, S. et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 27, 1247–1259 (2013).
Scharner, J. & Aznarez, I. Clinical applications of single-stranded oligonucleotides: current landscape of approved and in-development therapeutics. Mol. Ther. 29, 540–554 (2021).
Monani, U. R. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn–/– mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 9, 333–339 (2000).
Singh, N. K., Singh, N. N., Androphy, E. J. & Singh, R. N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 26, 1333–1346 (2006).
Hua, Y., Vickers, T. A., Okunola, H. L., Bennett, C. F. & Krainer, A. R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82, 834–848 (2008).
Hua, Y. et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126 (2011).
Hua, Y. et al. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 29, 288–297 (2015).
Chiriboga, C. A. et al. Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology 86, 890–897 (2016).
Rigo, F. et al. Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther. 350, 46–55 (2014).
Marasco, L. E. et al. Counteracting chromatin effects of a splicing-correcting antisense oligonucleotide improves its therapeutic efficacy in spinal muscular atrophy. Cell 185, 2057–2070 (2022).
Darras, B. T. et al. Risdiplam-treated infants with type 1 spinal muscular atrophy versus historical controls. N. Engl. J. Med. 385, 427–435 (2021).
Arechavala-Gomeza, V. et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 18, 798–810 (2007).
Aartsma-Rus, A. et al. Development of exon skipping therapies for Duchenne muscular dystrophy: a critical review and a perspective on the outstanding issues. Nucleic Acid. Ther. 27, 251–259 (2017).
Koenig, M. et al. The molecular basis for duchenne versus becker muscular dystrophy: correlation of severity with type of deletion. Tha Am. J. Hum. Genet. 45, 498–506 (1989).
Wright, C. J., Smith, C. W. J. & Jiggins, C. D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00514-4 (2022).
Cunningham, F. et al. Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019).
Lewis, B. P., Green, R. E. & Brenner, S. E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA 100, 189–192 (2003).
Kovalak, C., Donovan, S., Bicknell, A. A., Metkar, M. & Moore, M. J. Deep sequencing of pre-translational mRNPs reveals hidden flux through evolutionarily conserved alternative splicing nonsense-mediated decay pathways. Genome Biol. 22, 132 (2021).
Keren, H., Lev-Maor, G. & Ast, G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 11, 345–355 (2010).
Ashraf, U., Benoit-Pilven, C., Lacroix, V., Navratil, V. & Naffakh, N. Advances in analyzing virus-induced alterations of host cell splicing. Trends Microbiol. 27, 268–281 (2019).
Szakonyi, D. & Duque, P. Alternative splicing as a regulator of early plant development. Front. Plant. Sci. 9, 1174 (2018).
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A. & Brown, J. W. S. Alternative splicing in plants — coming of age. Trends Plant. Sci. 17, 616–623 (2012).
Godoy Herz, M. A. et al. Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73, 1066–1074 (2019).
Sanchez, S. E. et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 (2010).
Fairbrother, W. G., Yeh, R.-F., Sharp, P. A. & Burge, C. B. Predictive identification of exonic splicing enhancers in human genes. Science 297, 1007–1013 (2002).
Acknowledgements
The authors apologize to those researchers whose work could not be cited owing to space constraints. They thank B. Blencowe for critical reading of the manuscript and L. Giono for the design of Fig. 3B. Work in the authors’ laboratory was supported by a joint grant from Familias Atrofia Muscular Espinal (FAME, Argentina) and CureSMA (USA), and grants from the Lounsbery Foundation (USA), the Universidad de Buenos Aires (UBACYT 20020170100046BA), the Agencia Nacional de Promoción Científica y Tecnológica of Argentina (PICT-2019 862) and the CONICET (PUE 22920170100062CO).
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, made substantial contribution to discussion of content and reviewed and/or edited the manuscript before submission. A.R.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Jimena Giudice, Juan Valcárcel, and Leilei Chen, who co-reviewed with Han Jian, for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
National Human Genome Research Institute, National Institutes of Health (NIH): https://www.genome.gov/genetics-glossary/Exon
Glossary
- Transesterification
-
The exchange of the organic group R″ of an ester with the organic group R′ of an alcohol. In the second step of splicing, the uncleaved intron 3′ end is the (phosphodi)ester and the free exon is the alcohol. Following the reaction, the free intron becomes an alcohol and the joined exons form a phosphodiester bond.
- Cassette exons
-
Exons that as a whole are either included in or skipped from the mature mRNA.
- Class I exons
-
Exons whose inclusion in the mature mRNA is promoted by slow, and inhibited by fast, transcript elongation.
- Class II exons
-
Exons whose inclusion in the mature mRNA is inhibited by slow, and promoted by fast, transcript elongation.
- Nonsense-mediated decay
-
(NMD). An mRNA degradation mechanism that is coupled to splicing and translation and is triggered by premature stop codons.
- Detained introns
-
Introns that cause the mRNAs that harbour them to remain in the nucleus.
- γ-Aminobutyric acid A
-
(GABAA). A small organic molecule synthesized from the amino acid glutamic acid that acts as a neurotransmitter whose receptors are part of ligand-gated ion channels.
- Peptidergic secretion
-
The secretion of small polypeptides with neurotransmitter activity.
- Exon shuffling
-
A mechanism for the formation of new genes in eukaryotes during evolution, usually through recombination between introns of different genes, yielding novel rearranged genes with altered functions, without elimination of the original genes.
- r-Selection
-
A type of evolutionary selective pressure. r-Selected species, such as bacteria, are small organisms with short life cycles and fast maturation that are able to rapidly populate new environments.
- K-Selection
-
A type of evolutionary selective pressure. K-Selected species, such as pluricellular organisms with specialized tissues and organs, are bigger, have longer life cycles and slow maturation, and their colonization of new environments depends on their ability to adapt physiologically.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marasco, L.E., Kornblihtt, A.R. The physiology of alternative splicing. Nat Rev Mol Cell Biol 24, 242–254 (2023). https://doi.org/10.1038/s41580-022-00545-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-022-00545-z
This article is cited by
-
Loss-of-function mutation in PRMT9 causes abnormal synapse development by dysregulation of RNA alternative splicing
Nature Communications (2024)
-
Cellular zinc metabolism and zinc signaling: from biological functions to diseases and therapeutic targets
Signal Transduction and Targeted Therapy (2024)
-
On the genetic basis of tail-loss evolution in humans and apes
Nature (2024)
-
Alternative splicing and related RNA binding proteins in human health and disease
Signal Transduction and Targeted Therapy (2024)
-
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Nature Reviews Molecular Cell Biology (2024)