Abstract
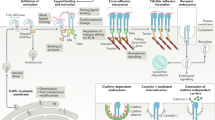
The ability of animal cells to sense, adhere to and remodel their local extracellular matrix (ECM) is central to control of cell shape, mechanical responsiveness, motility and signalling, and hence to development, tissue formation, wound healing and the immune response. Cell–ECM interactions occur at various specialized, multi-protein adhesion complexes that serve to physically link the ECM to the cytoskeleton and the intracellular signalling apparatus. This occurs predominantly via clustered transmembrane receptors of the integrin family. Here we review how the interplay of mechanical forces, biochemical signalling and molecular self-organization determines the composition, organization, mechanosensitivity and dynamics of these adhesions. Progress in the identification of core multi-protein modules within the adhesions and characterization of rearrangements of their components in response to force, together with advanced imaging approaches, has improved understanding of adhesion maturation and turnover and the relationships between adhesion structures and functions. Perturbations of adhesion contribute to a broad range of diseases and to age-related dysfunction, thus an improved understanding of their molecular nature may facilitate therapeutic intervention in these conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Humphrey, J. D., Dufresne, E. R. & Schwartz, M. A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014).
SenGupta, S., Parent, C. A. & Bear, J. E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 22, 529–547 (2021).
Romani, P., Valcarcel-Jimenez, L., Frezza, C. & Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 22, 22–38 (2021).
Cooper, J. & Giancotti, F. G. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347–367 (2019).
Bonnans, C., Chou, J. & Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801 (2014).
Kechagia, J. Z., Ivaska, J. & Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457–473 (2019).
Hayward, M. K., Muncie, J. M. & Weaver, V. M. Tissue mechanics in stem cell fate, development, and cancer. Dev. Cell 56, 1833–1847 (2021).
Hamidi, H. & Ivaska, J. Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018).
Chastney, M. R., Conway, J. R. W. & Ivaska, J. Integrin adhesion complexes. Curr. Biol. 31, R536–R542 (2021).
Bachmann, M., Kukkurainen, S., Hytonen, V. P. & Wehrle-Haller, B. Cell adhesion by integrins. Physiol. Rev. 99, 1655–1699 (2019).
Kadry, Y. A. & Calderwood, D. A. Chapter 22: Structural and signaling functions of integrins. Biochim. Biophys. Acta Biomembr. 1862, 183206 (2020).
Humphries, J. D., Chastney, M. R., Askari, J. A. & Humphries, M. J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 56, 14–21 (2019).
Slack, R. J., Macdonald, S. J. F., Roper, J. A., Jenkins, R. G. & Hatley, R. J. D. Emerging therapeutic opportunities for integrin inhibitors. Nat. Rev. Drug Discov. 21, 60–78 (2022).
Yamada, K. M., Doyle, A. D. & Lu, J. Cell–3D matrix interactions: recent advances and opportunities. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2022.03.002 (2022).
Sun, Z., Costell, M. & Fassler, R. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 21, 25–31 (2019).
Seetharaman, S. et al. Microtubules tune mechanosensitive cell responses. Nat. Mater. 21, 366–377 (2022).
Seetharaman, S. & Etienne-Manneville, S. Microtubules at focal adhesions — a double-edged sword. J. Cell Sci. https://doi.org/10.1242/jcs.232843 (2019).
Bouvard, D. et al. Functional consequences of integrin gene mutations in mice. Circ. Res. 89, 211–223 (2001).
Julich, D., Geisler, R. & Holley, S. A. Integrin α5 and Delta/Notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev. Cell 8, 575–586 (2005).
Williams, B. D. & Waterston, R. H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124, 475–490 (1994).
DeSimone, D. W., Dzamba, B. & Davidson, L. A. Using Xenopus embryos to investigate integrin function. Methods Enzymol. 426, 403–414 (2007).
Maartens, A. P. & Brown, N. H. Anchors and signals: the diverse roles of integrins in development. Curr. Top. Dev. Biol. 112, 233–272 (2015).
Luo, B. H., Carman, C. V. & Springer, T. A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 (2007).
Campbell, I. D. & Humphries, M. J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a004994 (2011).
Cormier, A. et al. Cryo-EM structure of the αvβ8 integrin reveals a mechanism for stabilizing integrin extension. Nat. Struct. Mol. Biol. 25, 698–704 (2018).
Campbell, M. G. et al. Cryo-EM reveals integrin-mediated TGF-β activation without release from latent TGF-β. Cell 180, 490–501.e16 (2020).
Schumacher, S. et al. Structural insights into integrin α5β1 opening by fibronectin ligand. Sci. Adv. https://doi.org/10.1126/sciadv.abe9716 (2021).
Nesic, D. et al. Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin αIIbβ3 for ligand binding. Blood Adv. 5, 1781–1790 (2021).
Sorrentino, S. et al. Structural analysis of receptors and actin polarity in platelet protrusions. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2105004118 (2021). This cryo-electron tomography study of endogenous platelet αIIbβ3 integrin in intact platelets reveals heterogeneity in integrin conformations and resolves a bent unbound integrin structure.
Moore, T. I., Aaron, J., Chew, T. L. & Springer, T. A. Measuring integrin conformational change on the cell surface with super-resolution microscopy. Cell Rep. 22, 1903–1912 (2018).
Li, J. et al. Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 36, 629–645 (2017).
Li, J. & Springer, T. A. Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl Acad. Sci. USA 114, 4685–4690 (2017).
Li, J., Yan, J. & Springer, T. A. Low-affinity integrin states have faster ligand-binding kinetics than the high-affinity state. eLife https://doi.org/10.7554/eLife.73359 (2021). This detailed analysis of ligand binding kinetics for α4β1 and α5β1 integrins reveals that low-affinity states are both more abundant on the cell surface and bind ligand faster than high-affinity states, suggesting that ligand binding may precede activation by inside-out signalling.
Mould, A. P. et al. Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J. Biol. Chem. 280, 4238–4246 (2005).
Kiema, T. et al. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21, 337–347 (2006).
Liu, J. et al. Structural mechanism of integrin inactivation by filamin. Nat. Struct. Mol. Biol. 22, 383–389 (2015).
Liu, W., Draheim, K. M., Zhang, R., Calderwood, D. A. & Boggon, T. J. Mechanism for KRIT1 release of ICAP1-mediated suppression of integrin activation. Mol. Cell 49, 719–729 (2013).
Rantala, J. K. et al. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat. Cell Biol. 13, 1315–1324 (2011).
Calderwood, D. A., Campbell, I. D. & Critchley, D. R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013).
Tadokoro, S. et al. Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 (2003).
Lagarrigue, F., Kim, C. & Ginsberg, M. H. The Rap1–RIAM–talin axis of integrin activation and blood cell function. Blood 128, 479–487 (2016).
Geiger, T. & Zaidel-Bar, R. Opening the floodgates: proteomics and the integrin adhesome. Curr. Opin. Cell Biol. 24, 562–568 (2012).
Whittaker, C. A. et al. The echinoderm adhesome. Dev. Biol. 300, 252–266 (2006).
Zaidel-Bar, R., Itzkovitz, S., Ma’ayan, A., Iyengar, R. & Geiger, B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9, 858–867 (2007).
Humphries, J. D. et al. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2, ra51 (2009).
Horton, E. R. et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 17, 1577–1587 (2015). This work presents proteomic analysis of the IAC adhesome in mouse fibroblasts and computational integration with multiple other published adhesome data sets to generate a consensus integrin adhesome.
Atkinson, S. J. et al. The β3-integrin endothelial adhesome regulates microtubule-dependent cell migration. EMBO Rep. https://doi.org/10.15252/embr.201744578 (2018).
Robertson, J. et al. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat. Commun. 6, 6265 (2015).
Maddala, R. & Rao, P. V. Global phosphotyrosinylated protein profile of cell–matrix adhesion complexes of trabecular meshwork cells. Am. J. Physiol. Cell Physiol. 319, C288–C299 (2020).
Paradzik, M. et al. KANK2 links αVβ5 focal adhesions to microtubules and regulates sensitivity to microtubule poisons and cell migration. Front. Cell Dev. Biol. 8, 125 (2020).
Randles, M. J. et al. Basement membrane ligands initiate distinct signalling networks to direct cell shape. Matrix Biol. 90, 61–78 (2020).
Lock, J. G. et al. Reticular adhesions are a distinct class of cell–matrix adhesions that mediate attachment during mitosis. Nat. Cell Biol. 20, 1290–1302 (2018). This work uses microscopy and mass spectrometry proteomics to identify a new class of adhesion (see also Baschieri et al. 2018), here termed ‘reticular adhesions’, that persist through cell division.
Horton, E. R. et al. The integrin adhesome network at a glance. J. Cell Sci. 129, 4159–4163 (2016).
Dong, J. M. et al. Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal. 9, rs4 (2016).
Myllymaki, S. M. et al. Assembly of the β4-integrin interactome based on proximal biotinylation in the presence and absence of heterodimerization. Mol. Cell Proteom. 18, 277–293 (2019).
Te Molder, L., Hoekman, L., Kreft, M., Bleijerveld, O. & Sonnenberg, A. Comparative interactomics analysis reveals potential regulators of α6β4 distribution in keratinocytes. Biol. Open https://doi.org/10.1242/bio.054155 (2020).
Chastney, M. R. et al. Topological features of integrin adhesion complexes revealed by multiplexed proximity biotinylation. J. Cell Biol. https://doi.org/10.1083/jcb.202003038 (2020). This work uses proximity biotinylation with 16 IAC protein baits to generate a proximity-dependent adhesome, allowing interrogation of the proximal relationships between adhesome components and providing insights into the architecture of IACs.
Mekhdjian, A. H. et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell 28, 1467–1488 (2017).
Rahikainen, R., Ohman, T., Turkki, P., Varjosalo, M. & Hytonen, V. P. Talin-mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J. Cell Sci. https://doi.org/10.1242/jcs.226514 (2019).
Bouchet, B. P. et al. Talin–KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife https://doi.org/10.7554/eLife.18124 (2016).
Sun, Z. et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 18, 941–953 (2016).
Rafiq, N. B. M. et al. A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat. Mater. 18, 638–649 (2019).
Choi, C. K. et al. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 (2008).
Bertocchi, C., Goh, W. I., Zhang, Z. & Kanchanawong, P. Nanoscale imaging by superresolution fluorescence microscopy and its emerging applications in biomedical research. Crit. Rev. Biomed. Eng. 41, 281–308 (2013).
Case, L. B. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17, 880–892 (2015).
Kanchanawong, P. et al. Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010).
Rossier, O. et al. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 14, 1057–1067 (2012).
Spiess, M. et al. Active and inactive β1 integrins segregate into distinct nanoclusters in focal adhesions. J. Cell Biol. 217, 1929–1940 (2018).
Bachir, A. I. et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 24, 1845–1853 (2014).
Cavalcanti-Adam, E. A. et al. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 92, 2964–2974 (2007).
Arnold, M. et al. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem 5, 383–388 (2004).
Yu, C.-h, Law, J. B. K., Suryana, M., Low, H. Y. & Sheetz, M. P. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl Acad. Sci. USA 108, 20585–20590 (2011).
Iskratsch, T. et al. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell 27, 545–559 (2013).
Kalappurakkal, J. M. et al. Integrin mechano-chemical signaling generates plasma membrane nanodomains that promote cell spreading. Cell 177, 1738–1756.e23 (2019).
Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017).
Changede, R., Xu, X., Margadant, F. & Sheetz, M. P. Nascent integrin adhesions form on all matrix rigidities after integrin activation. Dev. Cell 35, 614–621 (2015).
Morgan, M. R., Humphries, M. J. & Bass, M. D. Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 8, 957–969 (2007).
Powner, D., Kopp, P. M., Monkley, S. J., Critchley, D. R. & Berditchevski, F. Tetraspanin CD9 in cell migration. Biochem. Soc. Trans. 39, 563–567 (2011).
Yanez-Mo, M., Barreiro, O., Gordon-Alonso, M., Sala-Valdes, M. & Sanchez-Madrid, F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 (2009).
Ivaska, J. & Heino, J. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu. Rev. Cell Dev. Biol. 27, 291–320 (2011).
del Pozo, M. A. et al. Integrins regulate Rac targeting by internalization of membrane domains. Science 303, 839–842 (2004).
Hemler, M. E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005).
Changede, R., Cai, H., Wind, S. J. & Sheetz, M. P. Integrin nanoclusters can bridge thin matrix fibres to form cell–matrix adhesions. Nat. Mater. 18, 1366–1375 (2019). This work presents a reductionist experimental platform that probes IAC functions at the length scale of individual ECM fibres.
LaFlamme, S. E., Thomas, L. A., Yamada, S. S. & Yamada, K. M. Single subunit chimeric integrins as mimics and inhibitors of endogenous integrin functions in receptor localization, cell spreading and migration, and matrix assembly. J. Cell Biol. 126, 1287–1298 (1994).
Critchley, D. R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 38, 235–254 (2009).
Chen, W.-T. & Singer, S. J. Immunoelectron microscopic studies of the sites of cell–substratum and cell–cell contacts in cultured fibroblasts. J. Cell Biol. 95, 205–222 (1982).
Franz, C. M. & Muller, D. J. Analyzing focal adhesion structure by atomic force microscopy. J. Cell Sci. 118, 5315–5323 (2005).
Patla, I. et al. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat. Cell Biol. 12, 909–915 (2010).
Paszek, M. J. et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat. Methods 9, 825–827 (2012).
Shtengel, G. et al. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 123, 273–294 (2014).
Xia, S., Yim, E. K. F. & Kanchanawong, P. Molecular organization of integrin-based adhesion complexes in mouse embryonic stem cells. ACS Biomater. Sci. Eng. 5, 3828–3842 (2019).
Liu, J. et al. Talin determines the nanoscale architecture of focal adhesions. Proc. Natl Acad. Sci. USA 112, E4864–E4873 (2015).
Kanchanawong, P. & Waterman, C. M. Localization-based super-resolution imaging of cellular structures. Methods Mol. Biol. 1046, 59–84 (2013).
Stubb, A. et al. Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. Nat. Commun. 10, 4756 (2019).
Orré, T. et al. Molecular motion and tridimensional nanoscale localization of kindlin control integrin activation in focal adhesions. Nat. Commun. 12, 1–17 (2021).
Bouissou, A. et al. Podosome force generation machinery: a local balance between protrusion at the core and traction at the ring. ACS Nano 11, 4028–4040 (2017). This SRM study demonstrates that the podosome ring contains 3D nanoscale architecture comparable with FAs.
del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Barnett, S. F. & Kanchanawong, P. Visualizing the ‘backbone’of focal adhesions. Emerg. Top. Life Sci. 2, 677–680 (2018).
Klapholz, B. et al. Alternative mechanisms for talin to mediate integrin function. Curr. Biol. 25, 847–857 (2015). This work uses Drosophila mutants to show that talin is needed for all integrin adhesion events during development but that different processes require different talin domains and partners and that talin may be organized differently in IACs in different tissues.
Peng, X., Nelson, E. S., Maiers, J. L. & DeMali, K. A. New insights into vinculin function and regulation. Int. Rev. Cell Mol. Biol. 287, 191–231 (2011).
Xia, S. & Kanchanawong, P. Nanoscale mechanobiology of cell adhesions. Semin. Cell Dev. Biol. 71, 53–67 (2017).
Vogel, V. & Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006).
Iskratsch, T., Wolfenson, H. & Sheetz, M. P. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014).
Hoffman, B. D., Grashoff, C. & Schwartz, M. A. Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 (2011).
Gardel, M. L., Schneider, I. C., Aratyn-Schaus, Y. & Waterman, C. M. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26, 315–333 (2010).
Parsons, J. T., Horwitz, A. R. & Schwartz, M. A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 (2010).
Jaqaman, K., Galbraith, J. A., Davidson, M. W. & Galbraith, C. G. Changes in single-molecule integrin dynamics linked to local cellular behavior. Mol. Biol. Cell 27, 1561–1569 (2016).
Paszek, M. J. et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319–325 (2014).
Tsunoyama, T. A. et al. Super-long single-molecule tracking reveals dynamic-anchorage-induced integrin function. Nat. Chem. Biol. 14, 497–506 (2018).
Shibata, A. C. et al. Rac1 recruitment to the archipelago structure of the focal adhesion through the fluid membrane as revealed by single-molecule analysis. Cytoskeleton 70, 161–177 (2013).
Kanchanawong, P. et al. Analysis of single integrin behavior in living cells. Biophys. J. 98, 558a (2010).
Liu, B. et al. Biosensors based on peptide exposure show single molecule conformations in live cells. Cell 184, 5670–5685.e23 (2021).
Friedrich, O. et al. Stretch in focus: 2D inplane cell stretch systems for studies of cardiac mechano-signaling. Front. Bioeng. Biotechnol. 7, 55 (2019).
Kamble, H., Barton, M. J., Jun, M., Park, S. & Nguyen, N. T. Cell stretching devices as research tools: engineering and biological considerations. Lab Chip 16, 3193–3203 (2016).
Strohmeyer, N., Bharadwaj, M., Costell, M., Fassler, R. & Muller, D. J. Fibronectin-bound α5β1 integrins sense load and signal to reinforce adhesion in less than a second. Nat. Mater. 16, 1262–1270 (2017). This single-cell force microscopy study demonstrates rapid and multi-phasic temporal response of IAC to mechanical force.
Massou, S. et al. Cell stretching is amplified by active actin remodelling to deform and recruit proteins in mechanosensitive structures. Nat. Cell Biol. 22, 1011–1023 (2020).
Murthy, S. E., Dubin, A. E. & Patapoutian, A. Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 18, 771–783 (2017).
Schwartz, M. A., Brown, E. J. & Fazeli, B. A 50-kDa integrin-associated protein is required for integrin-regulated calcium entry in endothelial cells. J. Biol. Chem. 268, 19931–19934 (1993).
Carragher, N. et al. Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene 25, 5726–5740 (2006).
Franco, S. J. et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6, 977–983 (2004).
Cortesio, C. L., Boateng, L. R., Piazza, T. M., Bennin, D. A. & Huttenlocher, A. Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J. Biol. Chem. 286, 9998–10006 (2011).
Chan, K. T., Bennin, D. A. & Huttenlocher, A. Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J. Biol. Chem. 285, 11418–11426 (2010).
Saxena, M., Changede, R., Hone, J., Wolfenson, H. & Sheetz, M. P. Force-induced calpain cleavage of talin is critical for growth, adhesion development, and rigidity sensing. Nano Lett. 17, 7242–7251 (2017).
Schwartz, M. A., Both, G. & Lechene, C. Effect of cell spreading on cytoplasmic pH in normal and transformed fibroblasts. Proc. Natl Acad. Sci. USA 86, 4525–4529 (1989).
Schwartz, M. A., Lechene, C. & Ingber, D. E. Insoluble fibronectin activates the Na/H antiporter by clustering and immobilizing integrin α5β1, independent of cell shape. Proc. Natl Acad. Sci. USA 88, 7849–7853 (1991).
Webb, B. A., Chimenti, M., Jacobson, M. P. & Barber, D. L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011).
Pedersen, S. & Counillon, L. The SLC9A-C mammalian Na+/H+ exchanger family: molecules, mechanisms, and physiology. Physiol. Rev. 99, 2015–2113 (2019).
Srivastava, J. et al. Structural model and functional significance of pH-dependent talin–actin binding for focal adhesion remodeling. Proc. Natl Acad. Sci. USA 105, 14436–14441 (2008).
Canales Coutino, B. & Mayor, R. Mechanosensitive ion channels in cell migration. Cell Dev. 166, 203683 (2021).
Kefauver, J. M., Ward, A. B. & Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576 (2020).
Jin, P., Jan, L. Y. & Jan, Y. N. Mechanosensitive ion channels: structural features relevant to mechanotransduction mechanisms. Annu. Rev. Neurosci. 43, 207–229 (2020).
Wolfenson, H., Yang, B. & Sheetz, M. P. Steps in mechanotransduction pathways that control cell morphology. Annu. Rev. Physiol. 81, 585–605 (2019).
Kong, F., Garcia, A. J., Mould, A. P., Humphries, M. J. & Zhu, C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009).
Yao, M. et al. The mechanical response of talin. Nat. Commun. 7, 11966 (2016). This work presents a comprehensive in vitro characterization of talin stretching and vinculin recruitment under mechanical force.
Vigouroux, C., Henriot, V. & Le Clainche, C. Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force. Nat. Commun. 11, 3116 (2020). This work presents a biochemical reconstitution approach which characterizes how mechanical forces control the association or dissociation of different binding partners to talin.
Le, S. et al. Mechanotransmission and mechanosensing of human α-actinin 1. Cell Rep. 21, 2714–2723 (2017).
Rognoni, L., Stigler, J., Pelz, B., Ylanne, J. & Rief, M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl Acad. Sci. USA 109, 19679–19684 (2012).
Atherton, P. et al. Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 6, 10038 (2015).
Margadant, F. et al. Mechanotransduction in vivo by repeated talin stretch–relaxation events depends upon vinculin. PLoS Biol. 9, e1001223 (2011).
Hu, X. et al. Cooperative vinculin binding to talin mapped by time resolved super resolution microscopy. Nano Lett. 16, 4062–4068 (2016).
Atherton, P. et al. Relief of talin autoinhibition triggers a force-independent association with vinculin. J. Cell Biol. https://doi.org/10.1083/jcb.201903134 (2020).
Han, S. J. et al. Pre-complexation of talin and vinculin without tension is required for efficient nascent adhesion maturation. eLife 10, e66151 (2021).
Brockman, J. M. et al. Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 15, 115–118 (2018).
Swaminathan, V. et al. Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl Acad. Sci. USA 114, 10648–10653 (2017).
Nordenfelt, P. et al. Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 8, 2047 (2017).
Tan, S. J. et al. Regulation and dynamics of force transmission at individual cell–matrix adhesion bonds. Sci. Adv. 6, eaax0317 (2020). This integrated single-molecule integrin force measurement and modelling study highlights the complexity of IAC mechanotransduction.
Chang, A. C. et al. Single molecule force measurements in living cells reveal a minimally tensioned integrin state. ACS Nano 10, 10745–10752 (2016).
Nordenfelt, P., Elliott, H. L. & Springer, T. A. Coordinated integrin activation by actin-dependent force during T-cell migration. Nat. Commun. 7, 13119 (2016).
Kumar, A. et al. Local tension on talin in focal adhesions correlates with F-actin alignment at the nanometer scale. Biophys. J. 115, 1569–1579 (2018).
Driscoll, T. P., Ahn, S. J., Huang, B., Kumar, A. & Schwartz, M. A. Actin flow-dependent and -independent force transmission through integrins. Proc. Natl Acad. Sci. USA 117, 32413–32422 (2020).
Kumar, A. et al. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213, 371–383 (2016).
Ringer, P. et al. Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1. Nat. Methods 14, 1090–1096 (2017).
Goult, B. T., Brown, N. H. & Schwartz, M. A. Talin in mechanotransduction and mechanomemory at a glance. J. Cell. Sci. https://doi.org/10.1242/jcs.258749 (2021).
Lemke, S. B., Weidemann, T., Cost, A. L., Grashoff, C. & Schnorrer, F. A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. PLoS Biol. 17, e3000057 (2019). This in vivo study uses a talin intramolecular tension sensor to probe mechanical force in Drosophila muscle during the course of embryonic development.
Hu, K., Ji, L., Applegate, K. T., Danuser, G. & Waterman-Storer, C. M. Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007).
Chan, C. E. & Odde, D. J. Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008).
Brown, C. M. et al. Probing the integrin–actin linkage using high-resolution protein velocity mapping. J. Cell Sci. 119, 5204–5214 (2006).
Elosegui-Artola, A., Trepat, X. & Roca-Cusachs, P. Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018).
Gardel, M. L. et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183, 999–1005 (2008).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Wu, Z., Plotnikov, S. V., Moalim, A. Y., Waterman, C. M. & Liu, J. Two distinct actin networks mediate traction oscillations to confer focal adhesion mechanosensing. Biophys. J. 112, 780–794 (2017).
Ponti, A., Machacek, M., Gupton, S. L., Waterman-Storer, C. M. & Danuser, G. Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 (2004).
Yu, M. et al. Force-dependent regulation of talin–KANK1 complex at focal adhesions. Nano Lett. 19, 5982–5990 (2019).
Pasapera, A. M., Schneider, I. C., Rericha, E., Schlaepfer, D. D. & Waterman, C. M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–890 (2010).
Lavelin, I. et al. Differential effect of actomyosin relaxation on the dynamic properties of focal adhesion proteins. PLoS ONE 8, e73549 (2013).
Stutchbury, B., Atherton, P., Tsang, R., Wang, D. Y. & Ballestrem, C. Distinct focal adhesion protein modules control different aspects of mechanotransduction. J. Cell Sci. 130, 1612–1624 (2017).
Hoffmann, J.-E., Fermin, Y., Stricker, R. L., Ickstadt, K. & Zamir, E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. eLife 3, e02257 (2014).
Dumbauld, D. W. et al. How vinculin regulates force transmission. Proc. Natl Acad. Sci. USA 110, 9788–9793 (2013).
Case, L. B., Ditlev, J. A. & Rosen, M. K. Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494 (2019).
Zhu, J. et al. GIT/PIX condensates are modular and ideal for distinct compartmentalized cell signaling. Mol. Cell 79, 782–796.e6 (2020).
Case, L. B., De Pasquale, M., Henry, L. & Rosen, M. K. Synergistic phase separation of two pathways promotes integrin clustering and nascent adhesion formation. eLife 11, e72588 (2022).
Wang, Y. et al. LIMD1 phase separation contributes to cellular mechanics and durotaxis by regulating focal adhesion dynamics in response to force. Dev. Cell 56, 1313–1325.e7 (2021).
Sun, X. et al. Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell 55, 468–482.e7 (2020).
Winkelman, J. D., Anderson, C. A., Suarez, C., Kovar, D. R. & Gardel, M. L. Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proc. Natl Acad. Sci. USA 117, 25532–25542 (2020).
Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell https://doi.org/10.1016/j.molcel.2022.05.018 (2022).
Musacchio, A. On the role of phase separation in the biogenesis of membraneless compartments. EMBO J. 41, e109952 (2022).
Ramella, M., Ribolla, L. M. & de Curtis, I. Liquid–liquid phase separation at the plasma membrane–cytosol interface: common players in adhesion, motility, and synaptic function. J. Mol. Biol. 434, 167228 (2022).
Moreno-Layseca, P., Icha, J., Hamidi, H. & Ivaska, J. Integrin trafficking in cells and tissues. Nat. Cell Biol. 21, 122–132 (2019).
Nolte, M. A., Nolte-‘t Hoen, E. N. M. & Margadant, C. Integrins control vesicular trafficking; new tricks for old dogs. Trends Biochem. Sci. 46, 124–137 (2021).
Huet-Calderwood, C. et al. Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 8, 570 (2017).
Fourriere, L. et al. RAB6 and microtubules restrict protein secretion to focal adhesions. J. Cell Biol. 218, 2215–2231 (2019).
Eisler, S. A. et al. A rho signaling network links microtubules to PKD controlled carrier transport to focal adhesions. eLife https://doi.org/10.7554/eLife.35907 (2018).
Nader, G. P., Ezratty, E. J. & Gundersen, G. G. FAK, talin and PIPKIγ regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 18, 491–503 (2016).
Stehbens, S. J. et al. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 (2014).
Chen, N. P., Sun, Z. & Fassler, R. The Kank family proteins in adhesion dynamics. Curr. Opin. Cell Biol. 54, 130–136 (2018).
Krylyshkina, O. et al. Nanometer targeting of microtubules to focal adhesions. J. Cell Biol. 161, 853–859 (2003).
Ezratty, E. J., Bertaux, C., Marcantonio, E. E. & Gundersen, G. G. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J. Cell Biol. 187, 733–747 (2009).
Ezratty, E. J., Partridge, M. A. & Gundersen, G. G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7, 581–590 (2005).
Paul, N. R., Jacquemet, G. & Caswell, P. T. Endocytic trafficking of integrins in cell migration. Curr. Biol. 25, R1092–R1105 (2015).
Moreno-Layseca, P. et al. Cargo-specific recruitment in clathrin- and dynamin-independent endocytosis. Nat. Cell Biol. 23, 1073–1084 (2021).
De Franceschi, N. et al. Selective integrin endocytosis is driven by interactions between the integrin α-chain and AP2. Nat. Struct. Mol. Biol. 23, 172–179 (2016).
Burckhardt, C. J., Minna, J. D. & Danuser, G. SH3BP4 promotes neuropilin-1 and α5-integrin endocytosis and is inhibited by Akt. Dev. Cell 56, 1164–1181.e12 (2021).
Yu, C. H. et al. Integrin-β3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 6, 8672 (2015).
Baschieri, F. et al. Frustrated endocytosis controls contractility-independent mechanotransduction at clathrin-coated structures. Nat. Commun. 9, 3825 (2018).
Zuidema, A. et al. Mechanisms of integrin αVβ5 clustering in flat clathrin lattices. J. Cell Sci. https://doi.org/10.1242/jcs.221317 (2018).
Lock, J. G. et al. Clathrin-containing adhesion complexes. J. Cell Biol. 218, 2086–2095 (2019).
Alfonzo-Mendez, M. A., Sochacki, K. A., Strub, M. P. & Taraska, J. W. Dual clathrin and integrin signaling systems regulate growth factor receptor activation. Nat. Commun. 13, 905 (2022).
Elkhatib, N. et al. Tubular clathrin/AP-2 lattices pinch collagen fibers to support 3D cell migration. Science https://doi.org/10.1126/science.aal4713 (2017).
Litschel, T. et al. Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 12, 2254 (2021).
Kelley, C. F. et al. Phosphoinositides regulate force-independent interactions between talin, vinculin, and actin. eLife https://doi.org/10.7554/eLife.56110 (2020).
Ciobanasu, C., Faivre, B. & Le Clainche, C. Actomyosin-dependent formation of the mechanosensitive talin–vinculin complex reinforces actin anchoring. Nat. Commun. 5, 3095 (2014).
Xu, C. S. et al. An open-access volume electron microscopy atlas of whole cells and tissues. Nature 599, 147–151 (2021).
Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science https://doi.org/10.1126/science.aaz5357 (2020).
Elosegui-Artola, A. et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016).
Oria, R. et al. Force loading explains spatial sensing of ligands by cells. Nature 552, 219–224 (2017). This work presents the refinement and application of the molecular clutch model to probe IAC sensing of ligand geometry.
Young, L. E. & Higgs, H. N. Focal adhesions undergo longitudinal splitting into fixed-width units. Curr. Biol. 28, 2033–2045.e5 (2018).
Hu, S. et al. Structured illumination microscopy reveals focal adhesions are composed of linear subunits. Cytoskeleton 72, 235–245 (2015).
Case, L. B. & Waterman, C. M. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015).
Te Molder, L., de Pereda, J. M. & Sonnenberg, A. Regulation of hemidesmosome dynamics and cell signaling by integrin α6β4. J. Cell Sci. https://doi.org/10.1242/jcs.259004 (2021).
Zuidema, A., Wang, W. & Sonnenberg, A. Crosstalk between cell adhesion complexes in regulation of mechanotransduction. Bioessays 42, e2000119 (2020).
Jacquemet, G. et al. Filopodome mapping identifies p130Cas as a mechanosensitive regulator of filopodia stability. Curr. Biol. 29, 202–216.e7 (2019).
Miihkinen, M. et al. Myosin-X and talin modulate integrin activity at filopodia tips. Cell Rep. 36, 109716 (2021).
Kusumi, A., Tsunoyama, T. A., Hirosawa, K. M., Kasai, R. S. & Fujiwara, T. K. Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 (2014).
Manley, S. et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008).
Chen, Y., Pasapera, A. M., Koretsky, A. P. & Waterman, C. M. Orientation-specific responses to sustained uniaxial stretching in focal adhesion growth and turnover. Proc. Natl Acad. Sci. USA 110, E2352–E2361 (2013).
Polacheck, W. J. & Chen, C. S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423 (2016).
Colin-York, H. et al. Super-resolved traction force microscopy (STFM). Nano Lett. 16, 2633–2638 (2016).
Brockman, J. M. et al. Live-cell super-resolved PAINT imaging of piconewton cellular traction forces. Nat. Methods 17, 1018–1024 (2020).
Morimatsu, M., Mekhdjian, A. H., Adhikari, A. S. & Dunn, A. R. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 13, 3985–3989 (2013).
Wang, X. & Ha, T. Defining single molecular forces required to activate integrin and Notch signaling. Science 340, 991–994 (2013).
Fischer, L. S., Rangarajan, S., Sadhanasatish, T. & Grashoff, C. Molecular force measurement with tension sensors. Annu. Rev. Biophys. 50, 595–616 (2021).
Gingras, A. R. et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 27, 458–469 (2008).
Lu, F. et al. Mechanism of integrin activation by talin and its cooperation with kindlin. Nat. Commun. 13, 2362 (2022).
Bu, W., Levitskaya, Z., Tan, S. M. & Gao, Y. G. Emerging evidence for kindlin oligomerization and its role in regulating kindlin function. J. Cell Sci. https://doi.org/10.1242/jcs.256115 (2021).
Kadry, Y. A., Maisuria, E. M., Huet-Calderwood, C. & Calderwood, D. A. Differences in self-association between kindlin-2 and kindlin-3 are associated with differential integrin binding. J. Biol. Chem. 295, 11161–11173 (2020).
Winograd-Katz, S. E., Fassler, R., Geiger, B. & Legate, K. R. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15, 273–288 (2014).
Das, J., Sharma, A., Jindal, A., Aggarwal, V. & Rawat, A. Leukocyte adhesion defect: where do we stand circa 2019? Genes Dis. 7, 107–114 (2020).
Alpha, K. M., Xu, W. & Turner, C. E. Paxillin family of focal adhesion adaptor proteins and regulation of cancer cell invasion. Int. Rev. Cell Mol. Biol. 355, 1–52 (2020).
Sulzmaier, F. J., Jean, C. & Schlaepfer, D. D. FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer 14, 598–610 (2014).
Nurden, P., Stritt, S., Favier, R. & Nurden, A. T. Inherited platelet diseases with normal platelet count: phenotypes, genotypes and diagnostic strategy. Haematologica 106, 337–350 (2021).
Heller, K. N. et al. Human α7 integrin gene (ITGA7) delivered by adeno-associated virus extends survival of severely affected dystrophin/utrophin-deficient mice. Hum. Gene Ther. 26, 647–656 (2015).
Has, C. & Fischer, J. Inherited epidermolysis bullosa: new diagnostics and new clinical phenotypes. Exp. Dermatol. 28, 1146–1152 (2019).
Sorio, C. et al. Mutations of cystic fibrosis transmembrane conductance regulator gene cause a monocyte-selective adhesion deficiency. Am. J. Respir. Crit. Care Med. 193, 1123–1133 (2016).
Iwamoto, D. V. et al. Structural basis of the filamin A actin-binding domain interaction with F-actin. Nat. Struct. Mol. Biol. 25, 918–927 (2018).
Henderson, N. C., Rieder, F. & Wynn, T. A. Fibrosis: from mechanisms to medicines. Nature 587, 555–566 (2020).
Kim, K. K., Sheppard, D. & Chapman, H. A. TGF-β1 signaling and tissue fibrosis. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a022293 (2018).
Dong, X. et al. Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59 (2017).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Jun, J. I. & Lau, L. F. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol. 12, 676–685 (2010).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Meyer, K., Hodwin, B., Ramanujam, D., Engelhardt, S. & Sarikas, A. Essential role for premature senescence of myofibroblasts in myocardial fibrosis. J. Am. Coll. Cardiol. 67, 2018–2028 (2016).
Cho, K. A. et al. Morphological adjustment of senescent cells by modulating caveolin-1 status. J. Biol. Chem. 279, 42270–42278 (2004).
Rapisarda, V. et al. Integrin β3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. 18, 2480–2493 (2017).
Cheung, T. M. et al. Endothelial cell senescence increases traction forces due to age-associated changes in the glycocalyx and SIRT1. Cell Mol. Bioeng. 8, 63–75 (2015).
Chala, N. et al. Mechanical fingerprint of senescence in endothelial cells. Nano Lett. 21, 4911–4920 (2021).
Shin, E. Y. et al. Integrin-mediated adhesions in regulation of cellular senescence. Sci. Adv. 6, eaay3909 (2020).
Kuo, J. C., Han, X., Hsiao, C. T., Yates, J. R. III & Waterman, C. M. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393 (2011).
Acknowledgements
P.K. acknowledges funding support from the Mechanobiology Institute intramural fund and the Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2019-T2-2-014) and Tier 3 (MOE-T3-2020-0001). D.A.C. acknowledges funding support from the National Institutes of Health (NIH) (R01GM134148 and R01GM138411).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Kenneth Yamada, Johanna Ivaska and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- RGD ligands
-
Amino acid motifs comprising tripeptide arginine, glycine and aspartate that serve as an integrin binding site in extracellular matrix (ECM) proteins such as fibronectin and vitronectin.
- Basement membrane
-
A sheet of extracellular matrix (ECM) rich in laminins and type IV collagen to which epithelial and endothelial layers adhere. The basement membrane (sometimes termed the basal lamina) forms a barrier between tissues and provides a structural support and important signalling cues to epithelial and endothelial cells.
- FERM domain
-
The band 4.1, ezrin, radixin and moesin domain is found in various cytoskeletal-associated proteins, and typically contains three subdomains (F1, F2 and F3) arranged in a cloverleaf formation.
- Conformation-specific monoclonal antibodies
-
Antibodies that recognize the specific conformation of target proteins and, thus, can serve as reporters for the protein activation state or to selectively stabilize certain conformational states; for example, antibodies that recognize activated α5β1 integrin.
- Phosphoinositides
-
Membrane phospholipids with phosphorylated inositol headgroups, for example PIP2 (phosphatidyl inositol 4,5-bisphosphate) and PIP3 (phosphatidyl inositol 3,4,5-trisphosphate), recognized by different protein domains and important in signalling, vesicular trafficking and cellular compartmentalization.
- Yeast two-hybrid screens
-
A genetic screening method that relies on the modular domain organization of transcription factors, designed to identify binary protein–protein interactions between a specific ‘bait’ protein and a library of ‘prey’ proteins. In yeast cells, the ‘bait’ protein fused to a transcription factor DNA-binding domain is co-expressed with the library of ‘prey’ proteins fused to a transcriptional activation domain such that when ‘bait’ and ‘prey’ proteins interact the transcription factor is reconstituted, driving expression of target genes enabling selection of interacting clones and identification of ‘prey’ by DNA sequencing.
- Mass spectrometry proteomics
-
An analytical technique allowing the identification of proteins in complex samples on the basis of the mass to charge ratios of charged fragments.
- Src
-
A family of multi-domain signalling and adaptor proteins containing a tyrosine kinase domain, an Src homology 2 (SH2) domain which binds phosphorylated tyrosine and an SH3 domain which binds the proline-rich peptide motif.
- p130Cas
-
(130-kDa Crk-associated substrate (Cas)). An important scaffold protein encoded by the BCAR1 (breast cancer anti-oestrogen resistance 1) gene. p130Cas contains numerous phosphorylation sites for Src-family kinases (SFK) and serine/threonine kinases, a proline-rich region, an Src homology 3 (SH3) domain and binding sites to numerous integrin adhesion complex (IAC) components such as focal adhesion kinase (FAK), paxillin and kindlin.
- Cortical microtubule stabilizing complex
-
(CMSC). A micrometre-sized, patch-like, multi-protein complex localized in proximity to focal adhesions (FAs) and containing LL5β, liprins and ELKS adaptor proteins. Responsible for tethering microtubules at the cell cortex and linking them to FAs via KANK.
- Micelle
-
A particle suspension usually formed by segregation of a hydrophobic moiety in a lipid-like molecule away from the aqueous phase. The dimensions of micelle particles can be precisely determined by the molecular structure of the lipid, thus close-packing of micelle particles can be applied to achieve patterning at the nanoscale.
- Supported lipid bilayers
-
An experimental platform using fluid biological membranes formed on glass substrates to facilitate imaging-based study of membrane-dependent processes.
- Fluorescence anisotropy
-
A measurement of the fluorescence emission intensity differences along different polarization axes which can be used to estimate molecular orientation.
- GPI-anchored proteins
-
Peripheral membrane proteins that are anchored to the extracellular leaflet of the plasma membrane via post-translational modification that conjugate the protein C terminus with a polysaccharide linkage to GPI membrane lipid.
- Lipid rafts
-
Relatively ordered lateral domains (10–200 nm) in the plasma membranes that are enriched in cholesterol and sphingolipids.
- Syndecan
-
A family of homodimeric transmembrane proteoglycans that serve as receptors to the extracellular matrix (ECM) and growth factors via their extracellular glycosaminoglycan chains, and which contain small cytoplasmic domains with interaction sites for Src and Fyn tyrosine kinases and the PDZ (Post-synaptic-density-95/disc large tumour suppressor/zonula occludens 1) protein–protein interaction domain.
- Tetraspanin
-
A family of transmembrane proteins containing four transmembrane helices that can be organized into tetraspanin-enriched microdomains (TEMs), a sub-micrometre plasma membrane domain enriched with tetraspanins and cholesterol as well as associated membrane proteins such as integrins, and which form direct protein–protein interactions with a wide range of proteins including multiple integrins.
- Immuno-electron microscopy
-
An electron microscopy technique for localizing proteins in ultrastructure, in which specific biomolecules are identified using antibodies conjugated with electron-dense nanoparticles.
- Atomic force microscopy
-
An imaging and micro-manipulation technique which is based on cantilever-mounted mechanical probe tips, whereby laser-based measurement of the cantilever deflection allows high-precision measurement of physical topography or mechanical properties of the specimens.
- Cryo-electron tomography
-
An imaging technique in which 3D ultrastructural organization of cryogenically frozen specimens is determined from computation reconstruction of a series of electron micrographs taken at various angles.
- VASP
-
(Vasodilator stimulated phosphoprotein). VASP or Ena and their homologues Mena and EVL (Ena/Vasp-like) are tetrameric proteins known to promote barbed-end actin polymerization. In addition to actin and associated proteins, binding partners of VASP include zyxin, vinculin, Abl kinase and Src kinase.
- α-Actinin
-
A family of actin-binding proteins containing four isoforms in mammals (α-actinin 1 to α-actinin 4) which form antiparallel dimers that crosslink actin filaments.
- Cryptic vinculin binding sites
-
An amphipathic α-helix with high binding affinity to the vinculin head domain that is sterically sequestered by a folded protein tertiary structure, but which can be exposed upon mechanically induced protein conformational changes.
- Pleckstrin homology domain
-
A protein domain with a binding site for phosphatidylinositol glycolipid which confers the targeting ability to various cellular membrane compartments.
- Glycocalyx
-
A carbohydrate-rich biopolymer matrix associated with the exterior plasma membrane that plays important roles in cellular protection, cell–cell signalling, adhesion and recognition.
- Arp2/3
-
A heteroheptameric protein complex that promotes the formation of branched actin networks through filamentous actin nucleation and branching activities.
- Formin
-
A family of proteins that promote processive actin polymerization leading to the linear elongation of filamentous actin; examples include diaphanous formins (mDia1–mDia3).
- Mechanically activated ion channels
-
Transmembrane protein pores whose opening for specific ion conductance directly depends upon mechanical force, and which include calcium channels such as the Piezo family, members of the transient receptor potential (TRP) superfamily such as TRPV4 and potassium channels such as TREK1, TREK2 and TRAAK. Additionally, certain ligand-gated or voltage-dependent ion channels, such as Kv1.1, Nav1.5 and Hv1, have been shown to be modulated by mechanical force.
- Calpain
-
A calcium-dependent protease that cleaves specific sites in numerous integrin adhesion complex (IAC) components such as talin, focal adhesion kinase (FAK) and paxillin.
- Proton antiporters
-
Integral membrane proteins that regulate intracellular pH by coupling the export of proton with the import of cations into cells. Examples include sodium–hydrogen antiporter 1 (NHE1).
- Catch and slip bond
-
An intermolecular interface whose bond strength (typically measured as bond lifetime) varies as a function of force. With higher forces, slip bonds become weaker (that is, shorter lifetime) and catch bonds become stronger (that is, longer lifetime).
- Actin retrograde flow
-
The motion of filamentous actin networks from the cell periphery inwards, which is observed in various adherent cells. Actin retrograde flow is generally considered to be driven by actin polymerization and influenced by plasma membrane tension and myosin II contractility.
- FRET
-
(Förster resonance energy transfer). A process by which the energy of the excited state of a donor molecule is transferred to a nearby (typically <10 nm distance) acceptor molecule, which can be measured by observing fluorescence emission of the acceptor in response to the fluorescence excitation of the donor. Due to its sensitivity to molecular-scale changes in donor–acceptor proximity, the FRET principle is used in the design of a wide variety of biosensors (such as tension sensors, protein conformation sensors and enzymatic activity sensors).
- Myotendinous junction
-
A contact zone between muscles and tendon consisting of highly involuted muscle cell plasma membrane and serving as the main force transmission interface.
- Molecular clutch
-
A physical model describing mechanical force transmission between cell surface receptors through the flexible adaptor (or clutch) molecules that dynamically connect to actin or other cytoskeletal networks.
- βPIX
-
A protein encoded by the ARHGEF7 gene, also known as p21-activated protein kinase exchange factor-β, which can interact with GIT dimers and activate Rac1 and Cdc42 through its GTP exchange factor activity.
- GIT
-
GIT1 and GIT2 are related ARF GTPase activating proteins which form a stable complex with βPIX and play important roles in regulating small GTPase signalling.
- LIM domain
-
A protein domain found in a family of integrin adhesion complex (IAC) proteins such as zyxin, paxillin and LIMD1 that is capable of selectively binding to actin filaments under mechanical tension, named after the LIN11, ISL1 and MEC3 proteins.
- Durotaxis
-
Directional cell migration along a gradient of extracellular matrix (ECM) rigidity, typically towards regions of higher stiffness.
- Intrinsically disordered regions
-
Polypeptide segments that lack ordered conformation. Intrinsically disordered regions play diverse and important roles in protein functions, including serving as flexible linkers between protein domains, forming ligand recognition sites and participating in liquid–liquid phase separation (LLPS).
- Frustrated endocytosis
-
A modification of the classical clathrin-mediated endocytosis where a mechanical obstruction prevents endocytic structures from forming an invagination, resulting in long-lived, clathrin-coated, plasma membrane structures.
- Fibrinogen
-
A blood clotting factor which is cleaved by thrombin to form fibrin, which forms blood clots.
- Dystrophin-associated glycoprotein complex
-
A multi-protein complex found in various tissues that supports cell adhesion to laminin and linkage to the cytoskeleton. α-Dystroglycan binds laminin and associates with the transmembrane protein β-dystroglycan which engages the intracellular actin-binding adaptor protein dystrophin. Mutations in the complex are associated with muscular dystrophies and cardiomyopathies.
- Cystic fibrosis transmembrane conductance regulator
-
(CFTR). A transmembrane glycoprotein functioning as a chloride channel. Mutations in the CFTR gene cause cystic fibrosis and leukocyte adhesion deficiency (LAD) type IV.
- Poikiloderma
-
A skin condition characterized by patches or hyperpigmentation or hypopigmentation associated with atrophy.
- Latency-associated peptide
-
(LAP). The heavily glycosylated disulfide-linked dimer of the pro-peptide of the transforming growth factor-β (TGFβ) cytokine that binds TGFβ, preventing its interaction with its receptors.
- Senescence
-
The proliferative arrest of cells during ageing or in response to various stimuli such as genome instability or DNA damage. Contributes to developmental morphogenesis, suppresses tumour formation and limits fibrosis, but in ageing cells reduces cellular functions and regenerative capacities.
- Reactive oxygen species
-
(ROS). By-products of oxidative respiration by mitochondria such as hydrogen peroxide that play important roles in normal cellular processes including cell signalling and migration. Excess ROS production gives rise to oxidative stress and damages cellular structure.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kanchanawong, P., Calderwood, D.A. Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions. Nat Rev Mol Cell Biol 24, 142–161 (2023). https://doi.org/10.1038/s41580-022-00531-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-022-00531-5
This article is cited by
-
Resistance of HNSCC cell models to pan-FGFR inhibition depends on the EMT phenotype associating with clinical outcome
Molecular Cancer (2024)
-
Label-free biosensing with singular-phase-enhanced lateral position shift based on atomically thin plasmonic nanomaterials
Light: Science & Applications (2024)
-
Dynamic encounters with red blood cells trigger splenic marginal zone B cell retention and function
Nature Immunology (2024)
-
Fibroblast and myofibroblast activation in normal tissue repair and fibrosis
Nature Reviews Molecular Cell Biology (2024)
-
Focal adhesions contain three specialized actin nanoscale layers
Nature Communications (2024)