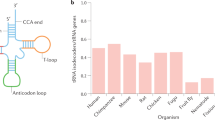
Abstract
Transfer RNA (tRNA) is an adapter molecule that links a specific codon in mRNA with its corresponding amino acid during protein synthesis. tRNAs are enzymatically modified post-transcriptionally. A wide variety of tRNA modifications are found in the tRNA anticodon, which are crucial for precise codon recognition and reading frame maintenance, thereby ensuring accurate and efficient protein synthesis. In addition, tRNA-body regions are also frequently modified and thus stabilized in the cell. Over the past two decades, 16 novel tRNA modifications were discovered in various organisms, and the chemical space of tRNA modification continues to expand. Recent studies have revealed that tRNA modifications can be dynamically altered in response to levels of cellular metabolites and environmental stresses. Importantly, we now understand that deficiencies in tRNA modification can have pathological consequences, which are termed ‘RNA modopathies’. Dysregulation of tRNA modification is involved in mitochondrial diseases, neurological disorders and cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Holley, R. W. et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965).
Crick, F. H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966).
Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Suzuki, T. in Fine-Tuning of RNA Functions by Modification and Editing, Vol. 12 (ed. Grosjean, H.) 23–69 (Springer-Verlag, 2005).
Agris, P. F. et al. Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15, 537–553 (2018).
Han, L. & Phizicky, E. M. A rationale for tRNA modification circuits in the anticodon loop. RNA 24, 1277–1284 (2018).
Bjork, G. R., Wikstrom, P. M. & Bystrom, A. S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244, 986–989 (1989).
Helm, M. & Alfonzo, J. D. Posttranscriptional RNA modifications: playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 21, 174–185 (2014).
Motorin, Y. & Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944 (2010).
Sylvers, L. A., Rogers, K. C., Shimizu, M., Ohtsuka, E. & Soll, D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 32, 3836–3841 (1993).
Suzuki, T., Ueda, T. & Watanabe, K. The ‘polysemous’ codon–a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16, 1122–1134 (1997).
Voorhees, R. M. & Ramakrishnan, V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236 (2013).
Florentz, C. & Giege, R. History of tRNA research in Strasbourg. IUBMB Life 71, 1066–1087 (2019).
Ibba, M. & Soll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000).
McClain, W. H. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234, 257–280 (1993).
Muramatsu, T. et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181 (1988).
Senger, B., Auxilien, S., Englisch, U., Cramer, F. & Fasiolo, F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry 36, 8269–8275 (1997).
Thiaville, P. C. et al. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol. 98, 1199–1221 (2015).
Niimi, T. et al. Recognition of the anticodon loop of tRNAIle1 by isoleucyl-transfer RNA synthetase from Escherichia coli. Nucleosides Nucleotides Nucleic Acids 13, 1231–1237 (1994).
Putz, J., Florentz, C., Benseler, F. & Giege, R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1, 580–582 (1994).
Uhlenbeck, O. C. & Schrader, J. M. Evolutionary tuning impacts the design of bacterial tRNAs for the incorporation of unnatural amino acids by ribosomes. Curr. Opin. Chem. Biol. 46, 138–145 (2018).
LaRiviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 (2001).
Asahara, H. & Uhlenbeck, O. C. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry 44, 11254–11261 (2005).
Lorenz, C., Lunse, C. E. & Morl, M. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7, 35 (2017).
Zhou, D., Tanzawa, T., Lin, J. & Gagnon, M. G. Structural basis for ribosome recycling by RRF and tRNA. Nat. Struct. Mol. Biol. 27, 25–32 (2020).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Juhling, F. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2009).
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Alfonzo, J. D. Post-transcriptional modifications are very important after all. RNA Biol. 11, 1481–1482 (2014).
Grosjean, H. in Fine-Tuning of RNA Functions by Modification and Editing, Vol. 12 (ed. Grosjean, H.) 1–22 (Springer-Verlag, 2005).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Kutter, C. et al. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat. Genet. 43, 948–955 (2011).
Gogakos, T. et al. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Hoffmann, A. et al. Accurate mapping of tRNA reads. Bioinformatics 34, 1116–1124 (2018).
Pinkard, O., McFarland, S., Sweet, T. & Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11, 4104 (2020).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Wang, J., Toffano-Nioche, C., Lorieux, F., Gautheret, D. & Lehmann, J. Accurate characterization of Escherichia coli tRNA modifications with a simple method of deep-sequencing library preparation. RNA Biol. https://doi.org/10.1080/15476286.2020.1790871 (2020).
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 (2016).
Song, J. et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 16, 160–169 (2020).
Kimura, S., Dedon, P. C. & Waldor, M. K. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 16, 964–972 (2020).
Suzuki, T., Nagao, A. & Suzuki, T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329 (2011).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020).
Marcus, J. O. & Bystrom, A. in Fine-Tuning of RNA Functions by Modification and Editing, Vol. 12 (ed. Grosjean, H.) 87–120 (Springer-Verlag, 2005).
Yoshida, M. et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl Acad. Sci. USA 112, 2764–2769 (2015).
Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589 (2002).
Asano, K. et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 46, 1565–1583 (2018).
Yokoyama, S. et al. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl Acad. Sci. USA 82, 4905–4909 (1985).
Kurata, S. et al. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U·G wobble pairing during decoding. J. Biol. Chem. 283, 18801–18811 (2008).
Kirino, Y. et al. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl Acad. Sci. USA 101, 15070–15075 (2004).
Johansson, M. J., Esberg, A., Huang, B., Bjork, G. R. & Bystrom, A. S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 28, 3301–3312 (2008).
Yasukawa, T., Suzuki, T., Ishii, N., Ohta, S. & Watanabe, K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20, 4794–4802 (2001).
Tukenmez, H., Xu, H., Esberg, A. & Bystrom, A. S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 43, 9489–9499 (2015).
Murphy, F. V. T., Ramakrishnan, V., Malkiewicz, A. & Agris, P. F. The role of modifications in codon discrimination by tRNALysUUU. Nat. Struct. Mol. Biol. 11, 1186–1191 (2004).
Zinshteyn, B. & Gilbert, W. V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 9, e1003675 (2013).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015).
Phelps, S. S., Malkiewicz, A., Agris, P. F. & Joseph, S. Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol. 338, 439–444 (2004).
Ranjan, N. & Rodnina, M. V. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J. Am. Chem. Soc. 139, 5857–5864 (2017).
Murphy, F. V. T. & Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 11, 1251–1252 (2004).
Wolf, J., Gerber, A. P. & Keller, W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 21, 3841–3851 (2002).
Gerber, A. P. & Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149 (1999).
Torres, A. G. et al. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 43, 5145–5157 (2015).
Novoa, E. M., Pavon-Eternod, M., Pan, T. & Ribas de Pouplana, L. A role for tRNA modifications in genome structure and codon usage. Cell 149, 202–213 (2012).
Moriya, J. et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry 33, 2234–2239 (1994).
Takemoto, C. et al. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 37, 1616–1627 (2009).
Cantara, W. A., Murphy, F. V. T., Demirci, H. & Agris, P. F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl Acad. Sci. USA 110, 10964–10969 (2013).
Nakano, S. et al. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol. 12, 546–551 (2016).
Van Haute, L. et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 7, 12039 (2016).
Haag, S. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 35, 2104–2119 (2016).
Kawarada, L. et al. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 45, 7401–7415 (2017).
Copela, L. A., Fernandez, C. F., Sherrer, R. L. & Wolin, S. L. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14, 1214–1227 (2008).
Kadaba, S. et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240 (2004).
Alexandrov, A. et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006).
Dewe, J. M., Whipple, J. M., Chernyakov, I., Jaramillo, L. N. & Phizicky, E. M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 18, 1886–1896 (2012).
Suzuki, T., Nagano, T. & Suzuki, T. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip. Rev. RNA 2, 376–386 (2011).
Umeda, N. et al. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 280, 1613–1624 (2005).
Huxtable, R. J. Physiological actions of taurine. Physiol. Rev. 72, 101–163 (1992).
Bouckenooghe, T., Remacle, C. & Reusens, B. Is taurine a functional nutrient? Curr. Opin. Clin. Nutr. Metab. Care 9, 728–733 (2006).
Verbrugghe, A. & Bakovic, M. Peculiarities of one-carbon metabolism in the strict carnivorous cat and the role in feline hepatic lipidosis. Nutrients 5, 2811–2835 (2013).
Morris, J. G. Idiosyncratic nutrient requirements of cats appear to be diet-induced evolutionary adaptations. Nutr. Res. Rev. 15, 153–168 (2002).
Schaffer, S. W., Jong, C. J., Warner, D., Ito, T. & Azuma, J. Taurine deficiency and MELAS are closely related syndromes. Adv. Exp. Med. Biol. 776, 153–165 (2013).
Sturman, J. A. Taurine in development. Physiol. Rev. 73, 119–147 (1993).
Seikai, T., Takeuchi, T. & Park, G. S. Comparison of growth, feed efficiency, and chemical composition of juvenile flounder fed live mysids and formula feed under laboratory conditions. Fish. Sci. 63, 520–526 (1997).
Sochacka, E. et al. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms - a factor important for accuracy of reading of guanosine at the 3-end of the mRNA codons. Nucleic Acids Res. 45, 4825–4836 (2017).
Kikuchi, G., Motokawa, Y., Yoshida, T. & Hiraga, K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84, 246–263 (2008).
Ducker, G. S. & Rabinowitz, J. D. One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017).
Morscher, R. J. et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554, 128–132 (2018).
Ye, J. et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4, 1406–1417 (2014).
Woo, C. C., Chen, W. C., Teo, X. Q., Radda, G. K. & Lee, P. T. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 7, 53005–53017 (2016).
Hashizume, O. et al. Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects. Sci. Rep. 5, 10434 (2015).
Molloy, A. M. Folate bioavailability and health. Int. J. Vitam. Nutr. Res. 72, 46–52 (2002).
Thiaville, P. C., Iwata-Reuyl, D. & de Crecy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 11, 1529–1539 (2014).
Schweizer, M. P., Chheda, G. B., Baczynskyj, L. & Hall, R. H. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. A new component of transfer ribonucleic acid. Biochemistry 8, 3283–3289 (1969).
Rozov, A., Demeshkina, N., Westhof, E., Yusupov, M. & Yusupova, G. Structural insights into the translational infidelity mechanism. Nat. Commun. 6, 7251 (2015).
Srinivasan, M. et al. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 30, 873–881 (2011).
El Yacoubi, B. et al. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 30, 882–893 (2011).
Lin, H. et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 9, 1875 (2018).
Smith, D. G., Pal, R. & Parker, D. Measuring equilibrium bicarbonate concentrations directly in cellular mitochondria and in human serum using europium/terbium emission intensity ratios. Chemistry 18, 11604–11613 (2012).
Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003).
Benej, M. et al. Carbonic anhydrase IX: regulation and role in cancer. Subcell. Biochem. 75, 199–219 (2014).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Nishimura, S. Structure, biosynthesis, and function of queuosine in transfer RNA. Prog. Nucleic Acid Res. Mol. Biol. 28, 49–73 (1983).
Kasai, H., Kuchino, Y., Nihei, K. & Nishimura, S. Distribution of the modified nucleoside Q and its derivatives in animal and plant transfer RNA’s. Nucleic Acids Res. 2, 1931–1939 (1975).
Iwata-Reuyl, D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg Chem. 31, 24–43 (2003).
Marks, T. & Farkas, W. R. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 230, 233–237 (1997).
Farkas, W. R. Effect of diet on the queuosine family of tRNAs of germ-free mice. J. Biol. Chem. 255, 6832–6835 (1980).
Hatfield, D. et al. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology 173, 736–742 (1989).
Muller, M. et al. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 47, 3711–3727 (2019).
Tuorto, F. et al. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777 (2018).
Iwata-Reuyl, D. & de Crecy-Lagard, V. in DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution, 377–391 (Landes Bioscience, 2009).
Chen, Y. C., Kelly, V. P., Stachura, S. V. & Garcia, G. A. Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure. RNA 16, 958–968 (2010).
Boland, C., Hayes, P., Santa-Maria, I., Nishimura, S. & Kelly, V. P. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J. Biol. Chem. 284, 18218–18227 (2009).
Rakovich, T. et al. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 286, 19354–19363 (2011).
Hanson, G. & Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016).
Wallace, D. C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 (2005).
Pavlakis, S. G., Phillips, P. C., DiMauro, S., De Vivo, D. C. & Rowland, L. P. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann. Neurol. 16, 481–488 (1984).
Fukuhara, N., Tokiguchi, S., Shirakawa, K. & Tsubaki, T. Myoclonus epilepsy associated with ragged-red fibres (mitochondrial abnormalities): disease entity or a syndrome? Light-and electron-microscopic studies of two cases and review of literature. J. Neurol. Sci. 47, 117–133 (1980).
Goto, Y., Nonaka, I. & Horai, S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 (1990).
Shoffner, J. M. et al. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell 61, 931–937 (1990).
Goto, Y., Nonaka, I. & Horai, S. A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS). Biochim. Biophys. Acta 1097, 238–240 (1991).
Kelley, S. O., Steinberg, S. V. & Schimmel, P. Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat. Struct. Biol. 7, 862–865 (2000).
Yasukawa, T. et al. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 275, 4251–4257 (2000).
Kirino, Y., Goto, Y., Campos, Y., Arenas, J. & Suzuki, T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl Acad. Sci. USA 102, 7127–7132 (2005).
Yasukawa, T. et al. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 579, 2948–2952 (2005).
Yasukawa, T. et al. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 467, 175–178 (2000).
Richter, U. et al. RNA modification landscape of the human mitochondrial tRNALys regulates protein synthesis. Nat. Commun. 9, 3966 (2018).
Hayashi, J. et al. Accumulation of mtDNA with a mutation at position 3271 in tRNALeu(UUR) gene introduced from a MELAS patient to HeLa cells lacking mtDNA results in progressive inhibition of mitochondrial respiratory function. Biochem. Biophys. Res. Commun. 197, 1049–1055 (1993).
Dunbar, D. R., Moonie, P. A., Zeviani, M. & Holt, I. J. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 5, 123–129 (1996).
Rikimaru, M. et al. Taurine ameliorates impaired the mitochondrial function and prevents stroke-like episodes in patients with MELAS. Intern. Med. 51, 3351–3357 (2012).
Ohsawa, Y. et al. Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicentre, open-label, 52-week phase III trial. J. Neurol. Neurosurg. Psychiatry 90, 529–536 (2019).
Torres, A. G., Batlle, E. & Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 20, 306–314 (2014).
Ramos, J. & Fu, D. The emerging impact of tRNA modifications in the brain and nervous system. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 412–428 (2019).
Ghezzi, D. et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 90, 1079–1087 (2012).
Baruffini, E. et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum. Mutat. 34, 1501–1509 (2013).
Fakruddin, M. et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 22, 482–496 (2018).
Kopajtich, R. et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 95, 708–720 (2014).
Zeharia, A. et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 85, 401–407 (2009).
Wu, Y. et al. Mtu1-mediated thiouridine formation of mitochondrial tRNAs is required for mitochondrial translation and is involved in reversible infantile liver injury. PLoS Genet. 12, e1006355 (2016).
Paramasivam, A., Meena, A. K., Venkatapathi, C., Pitceathly, R. D. S. & Thangaraj, K. Novel biallelic NSUN3 variants cause early-onset mitochondrial encephalomyopathy and seizures. J. Mol. Neurosci. 70, 1962–1965 (2020).
Patton, J. R., Bykhovskaya, Y., Mengesha, E., Bertolotto, C. & Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 280, 19823–19828 (2005).
Mangum, J. E. et al. Pseudouridine synthase 1 deficient mice, a model for mitochondrial myopathy with sideroblastic anemia, exhibit muscle morphology and physiology alterations. Sci. Rep. 6, 26202 (2016).
Carlile, T. M. et al. mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 15, 966–974 (2019).
Zhao, X. et al. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 15, 549–558 (2004).
Karijolich, J., Yi, C. & Yu, Y. T. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 16, 581–585 (2015).
Riley, C. M. et al. Central autonomic dysfunction with defective lacrimation; report of five cases. Pediatrics 3, 468–478 (1949).
Norcliffe-Kaufmann, L., Slaugenhaupt, S. A. & Kaufmann, H. Familial dysautonomia: history, genotype, phenotype and translational research. Prog. Neurobiol. 152, 131–148 (2017).
Anderson, S. L. et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68, 753–758 (2001).
Huang, B., Johansson, M. J. & Bystrom, A. S. An early step in wobble uridine tRNA modification requires the elongator complex. RNA 11, 424–436 (2005).
Johansson, M. J. O., Xu, F. & Bystrom, A. S. Elongator-a tRNA modifying complex that promotes efficient translational decoding. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 401–408 (2018).
Krutyholowa, R., Zakrzewski, K. & Glatt, S. Charging the code - tRNA modification complexes. Curr. Opin. Struct. Biol. 55, 138–146 (2019).
Karlsborn, T., Tukenmez, H., Chen, C. & Bystrom, A. S. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem. Biophys. Res. Commun. 454, 441–445 (2014).
Chen, Y. T. et al. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell. Biol. 29, 736–744 (2009).
Cohen, J. S. et al. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. A 167, 1391–1395 (2015).
Simpson, C. L. et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481 (2009).
Edvardson, S. et al. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 25, 545–551 (2017).
Braun, D. A. et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 49, 1529–1538 (2017).
Galloway, W. H. & Mowat, A. P. Congenital microcephaly with hiatus hernia and nephrotic syndrome in two sibs. J. Med. Genet. 5, 319–321 (1968).
Abbasi-Moheb, L. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 847–855 (2012).
Davarniya, B. et al. The role of a novel TRMT1 gene mutation and rare GRM1 gene defect in intellectual disability in two Azeri families. PLoS ONE 10, e0129631 (2015).
Freude, K. et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am. J. Hum. Genet. 75, 305–309 (2004).
Shaheen, R., Al-Salam, Z., El-Hattab, A. W. & Alkuraya, F. S. The syndrome dysmorphic facies, renal agenesis, ambiguous genitalia, microcephaly, polydactyly and lissencephaly (DREAM-PL): report of two additional patients. Am. J. Med. Genet. A 170, 3222–3226 (2016).
Alazami, A. M. et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J. Med. Genet. 50, 425–430 (2013).
Lentini, J. M., Alsaif, H. S., Faqeih, E., Alkuraya, F. S. & Fu, D. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat. Commun. 11, 2510 (2020).
Goodarzi, H. et al. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165, 1416–1427 (2016).
Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).
Pavon-Eternod, M. et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37, 7268–7280 (2009).
Santos, M., Fidalgo, A., Varanda, A. S., Oliveira, C. & Santos, M. A. S. tRNA deregulation and its consequences in cancer. Trends Mol. Med. 25, 853–865 (2019).
Delaunay, S. & Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 21, 552–559 (2019).
Shaheen, R. et al. Mutation in WDR4 impairs tRNA m7G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 16, 210 (2015).
Lin, S. et al. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255 e245 (2018).
Okamoto, M. et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 10, e1004639 (2014).
Gustavsson, M. & Ronne, H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 14, 666–674 (2008).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016).
Martinez, F. J. et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 49, 380–385 (2012).
Khan, M. A. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 856–863 (2012).
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014).
Rapino, F. et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609 (2018).
McMahon, M. & Ruggero, D. A wobbly road to drug resistance in melanoma: tRNA-modifying enzymes in translation reprogramming. EMBO J. 37, e99978 (2018).
Carlson, B. A. et al. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology 255, 2–8 (1999).
Waas, W. F., Druzina, Z., Hanan, M. & Schimmel, P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 282, 26026–26034 (2007).
Droogmans, L. & Grosjean, H. Enzymatic conversion of guanosine 3’ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J. 6, 477–483 (1987).
Noma, A., Kirino, Y., Ikeuchi, Y. & Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154 (2006).
Noma, A. et al. Expanding role of the jumonji C domain as an RNA hydroxylase. J. Biol. Chem. 285, 34503–34507 (2010).
Waas, W. F., de Crecy-Lagard, V. & Schimmel, P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J. Biol. Chem. 280, 37616–37622 (2005).
Grunberger, D., Weinstein, I. B. & Mushinski, J. F. Deficiency of the Y base in a hepatoma phenylalanine tRNA. Nature 253, 66–67 (1975).
Kuchino, Y., Borek, E., Grunberger, D., Mushinski, J. F. & Nishimura, S. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res. 10, 6421–6432 (1982).
Rossello-Tortella, M. et al. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc. Natl Acad. Sci. USA 117, 20785–20793 (2020).
Hatfield, D. et al. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology 173, 736–742 (1989).
Zhang, H. et al. GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin. Cancer Res. 20, 4717–4729 (2014).
Shen, Q. et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp. Cell Res. 315, 1653–1667 (2009).
Larrieu, D., Britton, S., Demir, M., Rodriguez, R. & Jackson, S. P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532 (2014).
Ito, S. et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18S ribosomal RNA (rRNA). J. Biol. Chem. 289, 35724–35730 (2014).
Sharma, S. et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258 (2015).
Hicks, D. G. et al. The expression of TRMT2A, a novel cell cycle regulated protein, identifies a subset of breast cancer patients with HER2 over-expression that are at an increased risk of recurrence. BMC Cancer 10, 108 (2010).
Fu, D. et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 30, 2449–2459 (2010).
Fu, Y. et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 49, 8885–8888 (2010).
Songe-Moller, L. et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 30, 1814–1827 (2010).
Shimada, K. et al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 69, 3157–3164 (2009).
Monies, D., Vagbo, C. B., Al-Owain, M., Alhomaidi, S. & Alkuraya, F. S. Recessive truncating mutations in ALKBH8 cause intellectual disability and severe impairment of wobble uridine modification. Am. J. Hum. Genet. 104, 1202–1209 (2019).
Steinthorsdottir, V. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 39, 770–775 (2007).
Arragain, S. et al. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 285, 28425–28433 (2010).
Wei, F. Y. et al. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121, 3598–3608 (2011).
Santos, M. et al. Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nat. Commun. 11, 296 (2020).
Suzuki, T., Ikeuchi, Y., Noma, A., Suzuki, T. & Sakaguchi, Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 425, 211–229 (2007).
Su, D. et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 9, 828–841 (2014).
Ross, R. L., Cao, X. & Limbach, P. A. Mapping post-transcriptional modifications onto transfer ribonucleic acid sequences by liquid chromatography tandem mass spectrometry. Biomolecules 7, 21 (2017).
Pomerantz, S. C. & McCloskey, J. A. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 193, 796–824 (1990).
Sakaguchi, Y., Miyauchi, K., Kang, B. I. & Suzuki, T. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 560, 19–28 (2015).
Heiss, M., Reichle, V. F. & Kellner, S. Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS. RNA Biol. 14, 1260–1268 (2017).
Ohira, T. & Suzuki, T. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol. 12, 648–655 (2016).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921 e910 (2020).
Smith, A. M., Jain, M., Mulroney, L., Garalde, D. R. & Akeson, M. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS ONE 14, e0216709 (2019).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216 e1226 (2018).
Yarham, J. W. et al. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 10, e1004424 (2014).
Powell, C. A. et al. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am. J. Hum. Genet. 97, 319–328 (2015).
Igoillo-Esteve, M. et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 9, e1003888 (2013).
Vilardo, E. & Rossmanith, W. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 43, 5112–5119 (2015).
Chen, J. & Patton, J. R. Pseudouridine synthase 3 from mouse modifies the anticodon loop of tRNA. Biochemistry 39, 12723–12730 (2000).
Lecointe, F. et al. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 273, 1316–1323 (1998).
Shaheen, R. et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 135, 707–713 (2016).
de Brouwer, A. P. M. et al. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 103, 1045–1052 (2018).
Arrondel, C. et al. Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 10, 3967 (2019).
Rodriguez, V. et al. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer 46, 694–707 (2007).
Rodriguez, V. et al. Structure-function analysis of human TYW2 enzyme required for the biosynthesis of a highly modified wybutosine (yW) base in phenylalanine-tRNA. PLoS ONE 7, e39297 (2012).
de Crecy-Lagard, V. et al. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res. 47, 2143–2159 (2019).
Miyauchi, K., Ohara, T. & Suzuki, T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 35, e24 (2007).
Suzuki, T. & Suzuki, T. Chaplet column chromatography: isolation of a large set of individual RNAs in a single step. Methods Enzymol. 425, 231–239 (2007).
Nagao, A. et al. Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria. Nat. Struct. Mol. Biol. 24, 778–782 (2017).
Krog, J. S. et al. 3-(3-amino-3-carboxypropyl)-5,6-dihydrouridine is one of two novel post-transcriptional modifications in tRNALys(UUU) from Trypanosoma brucei. FEBS J. 278, 4782–4796 (2011).
Miyauchi, K., Kimura, S. & Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 9, 105–111 (2013).
Matuszewski, M. et al. A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res. 45, 2137–2149 (2017).
Kang, B. I. et al. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 45, 2124–2136 (2017).
Salazar, J. C., Ambrogelly, A., Crain, P. F., McCloskey, J. A. & Soll, D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc. Natl Acad. Sci. USA 101, 7536–7541 (2004).
Ikeuchi, Y. et al. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6, 277–282 (2010).
Mandal, D. et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNAIle, base pairs with adenosine but not with guanosine. Proc. Natl Acad. Sci. USA 107, 2872–2877 (2010).
Mandal, D. et al. Identification and codon reading properties of 5-cyanomethyl uridine, a new modified nucleoside found in the anticodon wobble position of mutant haloarchaeal isoleucine tRNAs. RNA 20, 177–188 (2014).
Rose, S. et al. The hyperthermophilic partners Nanoarchaeum and Ignicoccus stabilize their tRNA T-loops via different but structurally equivalent modifications. Nucleic Acids Res. 48, 6906–6918 (2020).
Sakai, Y., Miyauchi, K., Kimura, S. & Suzuki, T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 44, 509–523 (2016).
Chen, P., Crain, P. F., Nasvall, S. J., Pomerantz, S. C. & Bjork, G. R. A “gain of function” mutation in a protein mediates production of novel modified nucleosides. EMBO J. 24, 1842–1851 (2005).
Dumelin, C. E., Chen, Y., Leconte, A. M., Chen, Y. G. & Liu, D. R. Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 8, 913–919 (2012).
Dal Magro, C. et al. A vastly increased chemical variety of RNA modifications containing a thioacetal structure. Angew. Chem. Int. Ed. 57, 7893–7897 (2018).
Jager, G., Chen, P. & Bjork, G. R. Transfer RNA bound to MnmH protein is enriched with geranylated tRNA–a possible intermediate in its selenation? PLoS ONE 11, e0153488 (2016).
Sierant, M. et al. Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 592, 2248–2258 (2018).
Reichle, V. F., Petrov, D. P., Weber, V., Jung, K. & Kellner, S. NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli. Nat. Commun. 10, 5600 (2019).
Acknowledgements
The author is grateful to members of his laboratory, in particular K. Miyauchi, T. Ohira and K. Minowa, for preparing tables and figures and for providing valuable suggestions. This work was supported by Grants-in-Aid for Scientific Research (18H05272) from JSPS, and Exploratory Research for Advanced Technology (ERATO; JPMJER2002) from JST. The author apologizes to those of his colleagues whose many contributions related to tRNA modifications could not be cited in this Review owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Peer review information
Nature Reviews Molecular Cell Biology thanks Juan Alfonzo, Tao Pan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Anticodon
-
A unit of three nucleotides at positions 34–36 in transfer RNA, which corresponds with three nucleotides of an mRNA codon.
- Reading frame maintenance
-
Of the three possible mRNA reading frames, only one reading frame generates the correct amino acid sequence. Transfer RNA modification in the anticodon is crucial for maintaining the correct reading frame.
- Acceptor stem
-
A stem structure made of seven base pairs formed by the 5′ and 3′ ends of transfer RNA. The 3′ CCA sequence and the discriminator base protrude from this stem.
- Discriminator base
-
The fourth nucleotide (position 73) from the 3′ end of transfer RNA (tRNA); frequently recognized by aminoacyl-tRNA synthetases and is a determinant of aminoacylation.
- Family box
-
In the genetic code, a codon box in which four codons are synonymous (encode the same amino acid).
- Two-codon sets
-
In the genetic code, a codon set in which two codons ending in pyrimidines or in purines are synonymous (specifying the same amino acid).
- Ribose puckering
-
A low-strain conformation of the ribose sugar ring in which atoms in the ring are displaced from the plane.
- C3′ endo form
-
Major puckering of ribose found in A-form duplex of RNA.
- Rapid tRNA decay
-
A pathway of 5′–3′ exonucleolytic degradation of mature transfer RNAs (tRNAs) that lack certain modifications.
- Warburg effect
-
Refers to tumours that metabolize glucose anaerobically rather than aerobically even when oxygen is available.
- Elongator complex
-
A transfer RNA-modifying enzyme complex catalysing 5-methoxyarbonylmethyluridine (mcm5U) formation.
Rights and permissions
About this article
Cite this article
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22, 375–392 (2021). https://doi.org/10.1038/s41580-021-00342-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-021-00342-0
This article is cited by
-
METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control
Experimental Hematology & Oncology (2024)
-
Selective gene expression maintains human tRNA anticodon pools during differentiation
Nature Cell Biology (2024)
-
RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives
Nature Reviews Gastroenterology & Hepatology (2024)
-
Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine
Nature Structural & Molecular Biology (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)