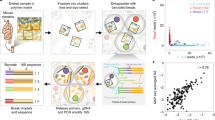
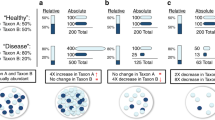
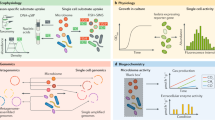
Abstract
Biogeography is the study of species distribution and diversity within an ecosystem and is at the core of how we understand ecosystem dynamics and interactions at the macroscale. In gut microbial communities, a historical reliance on bulk sequencing to probe community composition and dynamics has overlooked critical processes whereby microscale interactions affect systems-level microbiota function and the relationship with the host. In recent years, higher-resolution sequencing and novel single-cell level data have uncovered an incredible heterogeneity in microbial composition and have enabled a more nuanced spatial understanding of the gut microbiota. In an era when spatial transcriptomics and single-cell imaging and analysis have become key tools in mammalian cell and tissue biology, many of these techniques are now being applied to the microbiota. This fresh approach to intestinal biogeography has given important insights that span temporal and spatial scales, from the discovery of mucus encapsulation of the microbiota to the quantification of bacterial species throughout the gut. In this Review, we highlight emerging knowledge surrounding gut biogeography enabled by the observation and quantification of heterogeneity across multiple scales.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tropini, C. How the physical environment shapes the microbiota. mSystems 6, e0067521 (2021).
O’May, G. A., Reynolds, N., Smith, A. R., Kennedy, A. & Macfarlane, G. T. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J. Clin. Microbiol. 43, 3059–3065 (2005).
Ng, K. M. et al. Single-strain behavior predicts responses to environmental pH and osmolality in the gut microbiota. mBio 14, e00753-23 (2023).
Friedman, E. S. et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl Acad. Sci. USA 115, 4170–4175 (2018). The quantification of oxygen tension and microbial composition across gut regions reveals highly aerobic conditions in the proximal small instestine, and a corresponding high relative abundance of faculative anaerobes in the lumen and mucosa of this region.
Cremer, J., Arnoldini, M. & Hwa, T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc. Natl Acad. Sci. USA 114, 6438–6443 (2017).
Cremer, J. et al. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc. Natl Acad. Sci. USA 113, 11414–11419 (2016).
Vaishnava, S. et al. The antibacterial lectin regIIIg promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010).
Hansson, G. C. Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 15, 57–62 (2012).
Berry, D. et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl Acad. Sci. USA 110, 4720–4725 (2013).
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Tropini, C., Earle, K. A., Huang, K. C. & Sonnenburg, J. L. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21, 433–442 (2017).
Tropini, C. et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 173, 1742–1754.e17 (2018).
Peterson, D. A., Frank, D. N., Pace, N. R. & Gordon, J. I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 (2008).
Oren, A. & Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 71, 005056 (2021).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Manor, O. et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11, 5206 (2020).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Magne, F. et al. The Firmicutes/Bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12, 1474 (2020).
Moeller, A. H. The shrinking human gut microbiome. Curr. Opin. Microbiol. 38, 30–35 (2017).
Rausch, P. et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 306, 343–355 (2016).
Maghini, D. G. et al. Quantifying bias introduced by sample collection in relative and absolute microbiome measurements. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01754-3 (2023).
Jovel, J. et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7, 459 (2016).
Nearing, J. T. et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13, 342 (2022).
Schmidt, T. S. B. et al. Extensive transmission of microbes along the gastrointestinal tract. eLife 8, e42693 (2019).
Mcconnell, E. L., Basit, A. W. & Murdan, S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J. Pharm. Pharmacol. 60, 63–70 (2008).
Lkhagva, E. et al. The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol. 21, 44 (2021). A comprehensive profile of regional pH, water contents and microbiota composition across gastrointestinal regions provides a detailed reference for the gut biogeography of laboratory mice.
Quigley, E. M. M. & Turnberg, L. A. pH of the microclimate lining human gastric and duodenal mucosa in vivo studies in control subjects and in duodenal ulcer patients. Gastroenterology 92, 1876–1884 (1987).
Konradt, M. et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl Acad. Sci. USA 101, 5024–5029 (2004).
Stanforth, K. J. et al. Pepsin properties, structure, and its accurate measurement: a narrative review. Ann. Esophagus https://doi.org/10.21037/aoe-20-95 (2022).
Yeh Lee, Y., Erdogan, A. & Rao, S. S. C. How to assess regional and whole gut transit time with wireless motility capsule. J. Neurogastroenterol. Motil. 20, 265–270 (2014).
Hara, A. M. O. & Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 7, 688–693 (2006).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Bik, E. M. et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl Acad. Sci. USA 103, 732–737 (2006).
Wurm, P. et al. Qualitative and quantitative DNA- and RNA-based analysis of the bacterial stomach microbiota in humans, mice, and gerbils. mSystems 3, e00262-18 (2018).
Howitt, M. R. et al. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2, e00098-11 (2011).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
Li, X. et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE 4, e7985 (2009).
Sung, J. et al. Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J. Cancer Prev. 21, 60–65 (2016).
Evans, D. F. et al. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29, 1035–1041 (1988).
Johansson, M. E. V. & Hansson, G. C. Keeping bacteria at a distance. Science 334, 182–183 (2011).
Lueschow, S. R. & McElroy, S. J. The Paneth cell: the curator and defender of the immature small intestine. Front. Immunol. 11, 587 (2020).
Seekatz, A. M. et al. Spatial and temporal analysis of the stomach and small intestinal microbiota in fasted healthy humans. mSphere 4, e00126-19 (2019). A multi-channel catheter allows sampling of luminal microbiota composition and pH across the small intestine over time, revealing correlations between small intestine resident microorganism abundance and pH levels in individual participants.
Choi, C. H. & Chang, S. K. Role of small intestinal bacterial overgrowth in functional gastrointestinal disorders. J. Neurogastroenterol. Motil. 22, 3–5 (2016).
Husebye, E. The pathogenesis of gastrointestinal bacterial overgrowth. Chemotherapy 51, 1–22 (2005).
Könönen, E. & Gursoy, U. K. Oral Prevotella species and their connection to events of clinical relevance in gastrointestinal and respiratory tracts. Front. Microbiol. 12, 798763 (2022).
Tabula Sapiens Consortium. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022). Single-cell transcriptomics and bacterial sequencing across gut regions in human donors provides the first steps towards a high-resolution map of the microbial and transcriptional landscape of the human gut.
Sheth, R. U. et al. Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol. 37, 877–883 (2019). Plot-sampling methods usually reserved for macroscale ecosystems allows ~10–100-μm scale characterization of microbial community composition and organziation across gut regions.
Zoetendal, E. G. et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426 (2012).
Macfarlane, G. T., Gibson, G. R. & Cummings, J. H. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72, 57–64 (1992).
Johansson, M. E. V. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008).
Lee, K. S. et al. An automated Raman-based platform for the sorting of live cells by functional properties. Nat. Microbiol. 4, 1035–1048 (2019).
Luis, A. S. et al. A single sulfatase is required to access colonic mucin by a gut bacterium. Nature 598, 332–337 (2021).
Ge, X. et al. SRS-FISH: a high-throughput platform linking microbiome metabolism to identity at the single-cell level. Proc. Natl Acad. Sci. USA 119, e2203519119 (2022).
Gu, S. et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE 8, e74957 (2013).
James, K. R. et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353 (2020).
Pereira, F. C. & Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 19, 1366–1378 (2017).
Wexler, H. M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007).
Briliūtė, J. et al. Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat. Microbiol. 4, 1571–1581 (2019).
Donaldson, G. P. et al. Gut microbiota utilize immunoglobulin a for mucosal colonization. Science 360, 795–800 (2018). IgA is shown to interact with the surface capsule of B. fragilis to support colonization of the epithelium and crypts, and to differentially regulate epithelial colonization by other commensal bacteria in the colon.
Merchant, H. A., Liu, F., Orlu, M. & Basit, A. W. Age-mediated changes in the gastrointestinal tract. Int. J. Pharm. 512, 382–395 (2016).
Depner, M. et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat. Med. 26, 1766–1775 (2020).
Liu, Y. et al. Examination of the temporal and spatial dynamics of the gut microbiome in newborn piglets reveals distinct microbial communities in six intestinal segments. Sci. Rep. 9, 3453 (2019). Temporal sampling of regional microbiota composition from birth to adulthood in pigs provides insight into the dynamics of microbiota maturation in different gut compartments.
Mariat, D. et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9, 123 (2009).
Vaiserman, A. et al. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 20, 221 (2020).
Hopkins, M. J., Sharp, R. & Macfarlane, G. T. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48, 198–205 (2001).
Leite, G. et al. Age and the aging process significantly alter the small bowel microbiome. Cell Rep. 36, 109765 (2021).
Shalon, D. et al. Profiling the human intestinal environment under physiological conditions. Nature 617, 581–591 (2023).
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063.e8 (2014).
Espey, M. G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 55, 130–140 (2013).
Sun, W. W. et al. Nanoarchitecture and dynamics of the mouse enteric glycocalyx examined by freeze-etching electron tomography and intravital microscopy. Commun. Biol. 3, 5 (2020).
Layunta, E., Jäverfelt, S., Dolan, B., Arike, L. & Pelaseyed, T. IL-22 promotes the formation of a MUC17 glycocalyx barrier in the postnatal small intestine during weaning. Cell Rep. 34, 108757 (2021).
Huus, K. E., Petersen, C. & Finlay, B. B. Diversity and dynamism of IgA−microbiota interactions. Nat. Rev. Immunol. 21, 514–525 (2021).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Gassler, N. Paneth cells in intestinal physiology and pathophysiology. World J. Gastrointest. Pathophysiol. 8, 150–160 (2017).
Liu, Y. et al. The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression. Int. J. Biol. Macromol. 164, 884–891 (2020).
Birchenough, G. M. H., Nystrom, E. E. L., Johansson, M. E. V. & Hansson, G. C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016). A subpopulation of specialized crypt-resident goblet cells is characterized, which secrete mucus in response to contact with bacterial lipopolysaccharide to flush bacteria away from the crypt openings.
Kotarsky, K. et al. A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 3, 40–48 (2010).
Pédron, T. et al. A crypt-specific core microbiota resides in the mouse colon. mBio 3, e00116-12 (2012).
Pédron, T., Nigro, G. & Sansonetti, P. J. From homeostasis to pathology: decrypting microbe–host symbiotic signals in the intestinal crypt. Phil. Trans. R. Soc. B 371, 20150500 (2016).
Wang, W. et al. Three-dimensional quantitative imaging of native microbiota distribution in the gut. Angew. Chem. Int. Ed. Engl. 60, 3055–3061 (2021).
Saffarian, A. et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio 10, e01315-19 (2019).
Zaborin, A. et al. Spatial compartmentalization of the microbiome between the lumen and crypts is lost in the murine cecum following the process of surgery, including overnight fasting and exposure to antibiotics. mSystems https://doi.org/10.1128/msystems.00377-20 (2020).
Mondragón-Palomino, O. et al. Three-dimensional imaging for the quantification of spatial patterns in microbiota of the intestinal mucosa. Proc. Natl Acad. Sci. USA 119, e2118483119 (2022). Tissue clearing, 3D imaging and FISH staining allow high-resolution characterization of bacterial organization at the epithelium and within the crypts, and find large-scale disruptions to crypt microbiota architecture during and following antibiotic treatment.
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Dillon, A. & Lo, D. D. M cells: intelligent engineering of mucosal immune surveillance. Front. Immunol. 10, 1499 (2019).
Allaire, J. M. et al. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 39, 677–696 (2018).
Corr, S. C., Gahan, C. C. G. M. & Hill, C. M-cells: origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol. Med. Microbiol. 52, 2–12 (2008).
Vazquez-Torres, A. Cellular routes of invasion by enteropathogens. Curr. Opin. Microbiol. 3, 54–59 (2000).
Atarashi, K. et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015).
Ladinsky, M. S. et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363, eaat4042 (2019).
Ou, J., Liang, S., Guo, X. K. & Hu, X. α-Defensins promote Bacteroides colonization on mucosal reservoir to prevent antibiotic-induced dysbiosis. Front. Immunol. 11, 2065 (2020).
Vaga, S. et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci. Rep. 10, 14977 (2020).
Ndeh, D. et al. Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus. Nat. Commun. 11, 646 (2020).
Overbeeke, A. et al. Nutrient niche specificity for glycosaminoglycans is reflected in polysaccharide utilization locus architecture of gut Bacteroides species. Front. Microbiol. 13, 1033355 (2022).
Bergstrom, K. et al. Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science 370, 467–472 (2020). A careful analysis of the glycosylation patterns of mucus paired with regional mucin-knockout mice reveals that the outer mucus layer of the distal colon is proximal colon-derived, encapsulating the microbiota in transit through the lower intestine.
Nyström, E. E. L. et al. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 372, eabb1590 (2021).
Johansson, M. E. V., Holmén Larsson, J. M. & Hansson, G. C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc. Natl Acad. Sci. USA 108, 4659–4665 (2011).
Ermund, A., Schütte, A., Johansson, M. E. V., Gustafsson, J. K. & Hansson, G. C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 305, 341–347 (2013).
Szentkuti, L. & Lorenz, K. The thickness of the mucus layer in different segments of the rat intestine. Histochem. J. 27, 466–472 (1995).
Johansson, M. E. V. et al. Composition and functional role of the mucus layers in the intestine. Cell. Mol. Life Sci. 68, 3635–3641 (2011).
Duncan, K., Carey-Ewend, K. & Vaishnava, S. Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes 13, 1874815 (2021).
Johansson, M. E. V. et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291 (2014).
Tsai, H. H., Dwarakanath, A. D., Hart, C. A., Milton, J. D. & Rhodes, J. M. Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut 36, 570–576 (1995).
Krajina, B. A. et al. Dynamic light scattering microrheology reveals multiscale viscoelasticity of polymer gels and precious biological materials. ACS Cent. Sci. 16, 1294–1303 (2017).
Lieleg, O., Vladescu, I. & Ribbeck, K. Characterization of particle translocation through mucin hydrogels. Biophys. J. 98, 1782–1789 (2010).
Swidsinski, A. et al. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm. Bowel Dis. 13, 963–970 (2007).
Rogier, E. W., Frantz, A. L., Bruno, M. E. C. & Kaetzel, C. S. Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens 3, 390–403 (2014).
Barr, J. J. A bacteriophages journey through the human body. Immunol. Rev. 279, 106–122 (2017).
Zuppi, M., Hendrickson, H. L., O’Sullivan, J. M. & Vatanen, T. Phages in the gut ecosystem. Front. Cell. Infect. Microbiol. 11, 822562 (2022).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013).
Chin, W. H. et al. Bacteriophages evolve enhanced persistence to a mucosal surface. Proc. Natl Acad. Sci. USA 119, e2116197119 (2022).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401.e5 (2020).
Welch, J. L. M., Hasegawa, Y., McNulty, N. P., Gordon, J. I. & Borisy, G. G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017).
Jakobsson, H. E. et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177 (2015).
Li, H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015).
Yasuda, K. et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391 (2015).
Nava, G. M., Friedrichsen, H. J. & Stappenbeck, T. S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011).
Glover, J. S., Ticer, T. D. & Engevik, M. A. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci. Rep. 12, 8456 (2022).
Tailford, L. E., Crost, E. H., Kavanaugh, D. & Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81 (2015).
Derrien, M. et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2, 166 (2011).
Donaldson, G. P. et al. Spatially distinct physiology of Bacteroides fragilis within the proximal colon of gnotobiotic mice. Nat. Microbiol. 5, 746–756 (2020).
Engevik, M. A. et al. Bifidobacterium dentium fortifies the intestinal mucus layer. mBio 10, e01087-19 (2019).
Derrien, M., Belzer, C. & de Vos, W. M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 106, 171–181 (2017).
Chelakkot, C. et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 50, e450 (2018).
Ottman, N. et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 12, e0173004 (2017).
Hiippala, K. et al. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 10, 988 (2018).
Nagara, Y., Takada, T., Nagata, Y., Kado, S. & Kushiro, A. Microscale spatial analysis provides evidence for adhesive monopolization of dietary nutrients by specific intestinal bacteria. PLoS ONE 12, e0175497 (2017).
Shi, H. et al. Highly multiplexed spatial mapping of microbial communities. Nature 588, 676–681 (2020). Multiplexed FISH probes combined with spectral imaging enables the labelling and identification of up to 1,023 bacterial species within a sample and is used to characterize bacterial organization at the 10–100-μm scale in tissue samples before and after antibiotic treatment.
Macfarlane, S. & Macfarlane, G. T. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Environ. Microbiol. 72, 6204–6211 (2006).
Whitaker, W. R., Shepherd, E. S. & Sonnenburg, J. L. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169, 538–546.e12 (2017).
Li, D. et al. Microbial biogeography and core microbiota of the rat digestive tract. Sci. Rep. 8, 45840 (2017).
Mu, C., Yang, Y., Su, Y., Zoetendal, E. G. & Zhu, W. Differences in microbiota membership along the gastrointestinal tract of piglets and their differential alterations following an early-life antibiotic intervention. Front. Microbiol. 8, 797 (2017).
Teng, T. et al. Biogeography of the large intestinal mucosal and luminal microbiome in cynomolgus macaques with depressive-like behavior. Mol. Psychiatry 27, 1059–1067 (2022).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
De Cárcer, D. A. et al. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J. 5, 801–809 (2011).
Vijay, A. & Valdes, A. M. Challenges in nutrition role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 76, 489–501 (2022).
Nguyen, J., Pepin, D. M. & Tropini, C. Cause or effect? The spatial organization of pathogens and the gut microbiota in disease. Microbes Infect. 23, 104815 (2021).
Lennon, G. et al. Correlations between colonic crypt mucin chemotype, inflammatory grade and Desulfovibrio species in ulcerative colitis. Colorectal Dis. 16, 161–169 (2013).
Van Der Post, S. et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 68, 2142–2151 (2019). Biopsy explants from patients with ulcerative colitis allow the characterization of mucus layer integrity and production rate, and identify depletion of sentinel goblet cells as a mechanism of mucus layer thinning associated with IBD.
Hall, A. B. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). Extensive longitudinal sampling and metagenomic, metatransciptomic and metabolomic analysis of stool samples from patients with IBD and healthy controls link periods of dysbiosis and altered microbiome transcriptional activity to active disease.
Sugihara, K. et al. Mucolytic bacteria license pathobionts to acquire host-derived nutrients during dietary nutrient restriction. Cell Rep. 40, 111093 (2022).
Johansson, M. E. V. et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE 5, e12238 (2010).
Desai, M. S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). Dietary fibre is identified as a critical regulator of mucolytic bacteria abundance and colonic mucus barrier integrity; fibre-deprived mice develop thin, penetrable mucus layers, making them prone to colitis and pathogen infection.
Hamilton, M. K., Boudry, G., Lemay, D. G. & Raybould, H. E. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am. J. Physiol. Gastrointest. Live Physiol. 308, G840–G851 (2015).
Schroeder, B. O. et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe 23, 27–40.e7 (2018).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015).
Sheng, Q. S. et al. Comparison of gut microbiome in human colorectal cancer in paired tumor and adjacent normal tissues. Onco Targets Ther. 13, 635–646 (2020).
Burns, M. B., Lynch, J., Starr, T. K., Knights, D. & Blekhman, R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 7, 55 (2015).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Bi, D. et al. Profiling Fusobacterium infection at high taxonomic resolution reveals lineage-specific correlations in colorectal cancer. Nat. Commun. 13, 3336 (2022).
Han, Y. W. Fusobacterium nucleatum: a commensal-turned pathogen. Curr. Opin. Microbiol. 23, 141–147 (2015).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Yu, T. C. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 (2017).
Oliero, M. et al. Prevalence of pks + bacteria and enterotoxigenic Bacteroides fragilis in patients with colorectal cancer. Gut Pathog. 14, 51 (2022).
Ng, K. M. et al. Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe 26, 650–665.e4 (2019).
Poteres, E. et al. Selective regional alteration of the gut microbiota by diet and antibiotics. Front. Physiol. 11, 797 (2020).
Brown, C. T. et al. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2017).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Schmidt, F. et al. Noninvasive assessment of gut function using transcriptional recording sentinel cells. Science 376, eabm6038 (2022).
Riglar, D. T. et al. Bacterial variability in the mammalian gut captured by a single-cell synthetic oscillator. Nat. Commun. 10, 4665 (2019).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021).
Takahashi, H. et al. Direct detection of mRNA expression in microbial cells by fluorescence in situ hybridization using RNase H-assisted rolling circle amplification. Sci. Rep. 10, 9588 (2020).
Duboux, S., Muller, J. A., De Franceschi, F., Mercenier, A. & Kleerebezem, M. Using fluorescent promoter-reporters to study sugar utilization control in Bifidobacterium longum NCC 2705. Sci. Rep. 12, 10477 (2022).
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl Acad. Sci. USA 96, 9833–9838 (1999).
Geier, B. et al. Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nat. Microbiol. 5, 498–510 (2020).
Lin, L. et al. Revealing the in vivo growth and division patterns of mouse gut bacteria. Sci. Adv. 6, eabb2531 (2020).
Haugan, M. S., Charbon, G., Frimodt-Møller, N. & Løbner-Olesen, A. Chromosome replication as a measure of bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. Sci. Rep. 8, 14961 (2018).
Valm, A. M., Mark Welch, J. L. & Borisy, G. G. CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 35, 496–502 (2012).
Wilbert, S. A., Mark Welch, J. L. & Borisy, G. G. Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30, 4003–4015.e3 (2020).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Barr, J. J., Blackall, L. L. & Bond, P. Further limitations of phylogenetic group-specific probes used for detection of bacteria in environmental samples. ISME J. 4, 959–961 (2010).
Barrero-Canosa, J., Moraru, C., Zeugner, L., Fuchs, B. M. & Amann, R. Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ. Microbiol. 19, 70–82 (2017).
Shi, H., Grodner, B. & De Vlaminck, I. Recent advances in tools to map the microbiome. Curr. Opin. Biomed. Eng. 19, 100289 (2021).
Lloréns-Rico, V., Simcock, J. A., Huys, G. R. B. & Raes, J. Single-cell approaches in human microbiome research. Cell 185, 2725–2738 (2022).
Armetta, J. et al. Escherichia coli promoters with consistent expression throughout the murine gut. ACS Synth. Biol. 10, 3359–3368 (2021).
Nielsen, A. T. et al. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 6, e1001102 (2010).
Motta, J. P., Wallace, J. L., Buret, A. G., Deraison, C. & Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 18, 314–334 (2021).
Tytgat, H. L. P., Nobrega, F. L., van der Oost, J. & de Vos, W. M. Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 27, 17–25 (2019).
da Re, S. et al. Identification of commensal Escherichia coli genes involved in biofilm resistance to pathogen colonization. PLoS ONE 8, e61628 (2013).
Hassall, J., Cheng, J. K. J. & Unnikrishnan, M. Dissecting individual interactions between pathogenic and commensal bacteria within a multispecies gut microbial community. mSphere 6, e00013-21 (2021).
Baumgartner, M. et al. Mucosal biofilms are an endoscopic feature of irritable bowel syndrome and ulcerative colitis. Gastroenterology 161, 1245–1256.e20 (2021). Endoscopic, microscopic and genomic characterization of mucosal biofilms finds that differential localization and composition of biofilms are associated with IBD and left and right ulcerative colitis, and identifies common microbial species associated with each disease.
Mima, K. et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin. Transl Gastroenterol. 7, e200 (2016).
Dejea, C. M. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl Acad. Sci. USA 111, 18321–18326 (2014).
Kim, K., Castro, E. J. T., Shim, H., Advincula, J. V. G. & Kim, Y. W. Differences regarding the molecular features and gut microbiota between right and left colon cancer. Ann. Coloproctol. 34, 292–298 (2018).
Raskov, H., Kragh, K. N., Bjarnsholt, T., Alamili, M. & Gögenur, I. Bacterial biofilm formation inside colonic crypts may accelerate colorectal carcinogenesis. Clin. Transl Med. 7, 30 (2018).
Acknowledgements
The authors acknowledge that this work was completed on the traditional, ancestral and unceded territory of the xʷməθkʷəy̕əm (Musqueam) people. The authors encourage the reader to learn about the history of the land they work on at Native Land Digital (www.native-land.ca). They thank K. Ng, M. Orozco-Hidalgo, F. Papazotos, J. Burkhardt, H. Ghezzi and M. Hunter for their feedback and guidance in shaping this Review. G.M. acknowledges support from Natural Sciences and Engineering Research Council of Canada, Canada Graduate Scholarships.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Rustem Ismagilov, who co-reviewed with Ojas Pradhan and Natalie Woods, Alex Valm and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
McCallum, G., Tropini, C. The gut microbiota and its biogeography. Nat Rev Microbiol 22, 105–118 (2024). https://doi.org/10.1038/s41579-023-00969-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-023-00969-0
This article is cited by
-
Gut microbiota contributes to bisphenol A-induced maternal intestinal and placental apoptosis, oxidative stress, and fetal growth restriction in pregnant ewe model by regulating gut-placental axis
Microbiome (2024)
-
Gastrointestinal and brain barriers: unlocking gates of communication across the microbiota–gut–brain axis
Nature Reviews Gastroenterology & Hepatology (2024)
-
Does a Dysbiotic Oral Microbiome Trigger the Risk of Chronic Inflammatory Disease?
Current Treatment Options in Allergy (2023)