Abstract
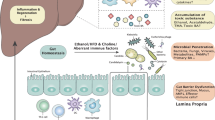
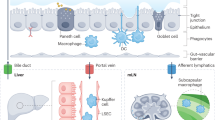
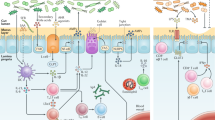
The trillions of microorganisms in the human intestine are important regulators of health, and disruptions in the gut microbial communities can cause disease. The gut, liver and immune system have a symbiotic relationship with these microorganisms. Environmental factors, such as high-fat diets and alcohol consumption, can disrupt and alter microbial communities. This dysbiosis can lead to dysfunction of the intestinal barrier, translocation of microbial components to the liver and development or progression of liver disease. Changes in metabolites produced by gut microorganisms can also contribute to liver disease. In this Review, we discuss the importance of the gut microbiota in maintenance of health and the alterations in microbial mediators that contribute to liver disease. We present strategies for modulation of the intestinal microbiota and/or their metabolites as potential treatments for liver disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mazagova, M. et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 29, 1043–1055 (2015).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012). To our knowledge, this paper is the first to report transmissibility of experimental steatohepatitis in mice by FMT.
Llopis, M. et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016). To our knowledge, this paper is the first to report transmissibility of ethanol-induced liver disease in gnotobiotic mice.
Philips, C. A., Ahamed, R., Rajesh, S., Abduljaleel, J. K. P. & Augustine, P. Long-term outcomes of stool transplant in alcohol-associated hepatitis-analysis of clinical outcomes, relapse, gut microbiota and comparisons with standard care. J. Clin. Exp. Hepatol. 12, 1124–1132 (2022).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Suhr, M. J., Banjara, N. & Hallen-Adams, H. E. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett. Appl. Microbiol. 62, 209–215 (2016).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71 (2021).
Hooper, L. V., Midtvedt, T. & Gordon, J. I. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 (2002).
Schulthess, J. et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 (2019).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Litvak, Y., Byndloss, M. X. & Baumler, A. J. Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018).
Lee, J. Y., Tsolis, R. M. & Baumler, A. J. The microbiome and gut homeostasis. Science 377, eabp9960 (2022).
Jakobsson, H. E. et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177 (2015).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008). This study demonstrates two distinct mucus layers coating the colonic epithelium, both comprised in large part by mucin 2, that separate bacteria from the colon epithelia.
Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Bunker, J. J. & Bendelac, A. IgA responses to microbiota. Immunity 49, 211–224 (2018).
Turner, J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809 (2009).
Bacher, P. et al. Human anti-fungal TH17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176, 1340–1355.e15 (2019).
McDonald, B. et al. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe 28, 660–668.e4 (2020).
Le, H. H., Lee, M. T., Besler, K. R. & Johnson, E. L. Host hepatic metabolism is modulated by gut microbiota-derived sphingolipids. Cell Host Microbe 30, 798–808.e7 (2022).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Moro-Sibilot, L. et al. Mouse and human liver contain immunoglobulin A-secreting cells originating from Peyer’s patches and directed against intestinal antigens. Gastroenterology 151, 311–323 (2016).
Inagaki, T. et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl Acad. Sci. USA 103, 3920–3925 (2006). This study elucidates the role of FXR in maintaining the intestinal barrier and inducing antimicrobial proteins, which prevents bacterial overgrowth and translocation.
Mouries, J. et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 71, 1216–1228 (2019).
Clements, W. D. et al. Role of the gut in the pathophysiology of extrahepatic biliary obstruction. Gut 39, 587–593 (1996).
Friedman, E. S. et al. FXR-dependent modulation of the human small intestinal microbiome by the bile acid derivative obeticholic acid. Gastroenterology 155, 1741–1752.e5 (2018).
Fuchs, C. D. & Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 19, 432–450 (2022).
Inagaki, T. et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 (2005).
Kliewer, S. A. & Mangelsdorf, D. J. Bile acids as hormones: the FXR–FGF15/19 pathway. Dig. Dis. 33, 327–331 (2015).
Powell, E. E., Wong, V. W. & Rinella, M. Non-alcoholic fatty liver disease. Lancet 397, 2212–2224 (2021).
Lee, G. et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 11, 4982 (2020).
Demir, M. et al. The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 76, 788–799 (2022).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–1062.e5 (2017).
Lang, S. et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159, 1839–1852 (2020). To our knowledge, this study is the first to interrogate the composition of the intestinal virome and its association with severity of NAFLD and fibrosis.
Rahman, K. et al. Loss of junctional adhesion molecule a promotes severe steatohepatitis in mice on a diet high in saturated fat, fructose, and cholesterol. Gastroenterology 151, 733–746.e12 (2016).
Hsu, C. L. et al. Differences in bacterial translocation and liver injury in ethanol versus diet-induced liver disease. Dig. Dis. Sci. https://doi.org/10.1007/s10620-023-07860-1 (2023).
Luther, J. et al. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell Mol. Gastroenterol. Hepatol. 1, 222–232 (2015).
Meijnikman, A. S. et al. Microbiome-derived ethanol in nonalcoholic fatty liver disease. Nat. Med. 28, 2100–2106 (2022). To our knowledge, this study is the first to measure portal vein ethanol concentrations in patients with NAFLD and demonstrate higher portal vein ethanol concentrations in patients post-prandially and with more advanced liver disease.
Zhu, L. et al. Characterization of the gut microbiome in non-alcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013).
Yuan, J. et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab. 30, 675–688.e7 (2019).
Hoyles, L. et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018).
Zhao, M. et al. TMAVA, a metabolite of intestinal microbes, is increased in plasma from patients with liver steatosis, inhibits γ-butyrobetaine hydroxylase, and exacerbates fatty liver in mice. Gastroenterology 158, 2266–2281.e27 (2020).
Beaumont, M. et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J. 32, fj201800544 (2018).
Ma, L. et al. Indole alleviates diet-induced hepatic steatosis and inflammation in a manner involving myeloid cell 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Hepatology 72, 1191–1203 (2020).
Teunis, C., Nieuwdorp, M. & Hanssen, N. Interactions between tryptophan metabolism, the gut microbiome and the immune system as potential drivers of non-alcoholic fatty liver disease (NAFLD) and metabolic diseases. Metabolites 12, 514 (2022).
Jiao, N. et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 67, 1881–1891 (2018).
Ferslew, B. C. et al. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig. Dis. Sci. 60, 3318–3328 (2015).
Mouzaki, M. et al. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS ONE 11, e0151829 (2016).
Fang, S. et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 21, 159–165 (2015).
Jiang, C. et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 125, 386–402 (2015).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019). This study identifies bile acid metabolites capable of regulating T cell differentiation, reducing TH17 cells and increasing Treg cells.
Brandl, K. et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 69, 396–405 (2018).
Li, J. & Dawson, P. A. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 895–911 (2019).
Hild, B. et al. Neonatal exposure to a wild-derived microbiome protects mice against diet-induced obesity. Nat. Metab. 3, 1042–1057 (2021).
Duan, Y. et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019). To our knowledge, this study is the first to demonstrate that phage-based therapy in a murine model of a non-infectious disease reduces ethanol-induced liver disease.
Grander, C. et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut 67, 891–901 (2018).
Chu, H. et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 72, 391–400 (2020).
Lang, S. et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology 71, 522–538 (2020). This study describes the faecal mycobiome in patients with alcohol-associated hepatitis.
Hsu, C. L. et al. Intestinal virome in patients with alcohol use disorder and after abstinence. Hepatol. Commun. 6, 2058–2069 (2022).
Hsu, C. L. et al. Any alcohol use in NAFLD patients is associated with significant changes to the intestinal virome. Hepatology 77, 2073–2083 (2023).
Jiang, L. et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology 72, 2182–2196 (2020).
Leclercq, S. et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl Acad. Sci. USA 111, E4485–E4493 (2014). To our knowledge, this study is the first demonstrating the association of gut permeability and gut-microbiota composition with addiction severity in patients with alcohol use disorder.
Lang, S. & Schnabl, B. Microbiota and fatty liver disease — the known, the unknown, and the future. Cell Host Microbe 28, 233–244 (2020).
Wang, L. et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 19, 227–239 (2016). This study demonstrates that intestinal antimicrobial molecules such as REG3 lectins protect the host from bacterial translocation to extra-intestinal tissues caused by chronic ethanol exposure.
Couch, R. D. et al. Alcohol induced alterations to the human fecal VOC metabolome. PLoS ONE 10, e0119362 (2015).
Cresci, G. A. et al. Prophylactic tributyrin treatment mitigates chronic-binge alcohol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol. 32, 1587–1597 (2017).
Morrison, D. J. & Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200 (2016).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 (2019).
Hartmann, P. et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology 67, 2150–2166 (2018).
Helsley, R. N. et al. Gut microbial trimethylamine is elevated in alcohol-associated hepatitis and contributes to ethanol-induced liver injury in mice. eLife 11, e76554 (2022).
Haas, W., Shepard, B. D. & Gilmore, M. S. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87 (2002).
Lang, S., Demir, M., Duan, Y., Martin, A. & Schnabl, B. Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver Int. 40, 860–865 (2020).
Yang, Y. et al. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature 607, 563–570 (2022).
Richardson, J. P. et al. Candidalysins are a new family of cytolytic fungal peptide toxins. mBio 13, e0351021 (2022).
Verma, A. H. et al. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci. Immunol. 2, eaam8834 (2017).
Kasper, L. et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat. Commun. 9, 4260 (2018).
Chang, C. S., Chen, G. H., Lien, H. C. & Yeh, H. Z. Small intestine dysmotility and bacterial overgrowth in cirrhotic patients with spontaneous bacterial peritonitis. Hepatology 28, 1187–1190 (1998).
Bauer, T. M. et al. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia. Am. J. Gastroenterol. 97, 2364–2370 (2002).
Bajaj, J. S. et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 70, 1162–1173 (2021).
Bajaj, J. S. et al. Fungal dysbiosis in cirrhosis. Gut 67, 1146–1154 (2018).
Hasa, E., Hartmann, P. & Schnabl, B. Liver cirrhosis and immune dysfunction. Int. Immunol. 34, 455–466 (2022).
Trebicka, J., Macnaughtan, J., Schnabl, B., Shawcross, D. L. & Bajaj, J. S. The microbiota in cirrhosis and its role in hepatic decompensation. J. Hepatol. 75, S67–S81 (2021).
Kakiyama, G. et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58, 949–955 (2013).
Sorribas, M. et al. FXR modulates the gut–vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J. Hepatol. 71, 1126–1140 (2019).
Kulik, L. & El-Serag, H. B. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 156, 477–491.e1 (2019).
Ren, Z. et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68, 1014–1023 (2019).
Ponziani, F. R. et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 69, 107–120 (2019).
Zheng, R. et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 9, 4232–4250 (2020).
Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Steck, N. et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971 (2011).
Iida, N. et al. Chronic liver disease enables gut Enterococcus faecalis colonization to promote liver carcinogenesis. Nat. Cancer 2, 1039–1054 (2021).
Yu, L. X. et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 52, 1322–1333 (2010).
Dapito, D. H. et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516 (2012).
Fedirko, V. et al. Exposure to bacterial products lipopolysaccharide and flagellin and hepatocellular carcinoma: a nested case–control study. BMC Med. 15, 72 (2017).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). To our knowledge, this study is the first to demonstrate that the microbiome of individuals with obesity has an increased capacity for energy harvest from the diet and that obesity was transmissible to germ-free mice with transplantation of this microbiota.
Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 1693, 128–133 (2018).
Braniste, V. et al. The gut microbiota influences blood–brain barrier permeability in mice. Sci. Transl Med. 6, 263ra158 (2014).
Bajaj, J. S. et al. A randomized clinical trial of fecal microbiota transplant for alcohol use disorder. Hepatology 73, 1688–1700 (2021). To our knowledge, this work is the first human study to demonstrate improvement in alcohol craving symptoms in patients with alcohol-associated cirrhosis and alcohol use disorder after FMT.
Bajaj, J. S. et al. Antibiotic-associated disruption of microbiota composition and function in cirrhosis is restored by fecal transplant. Hepatology 68, 1549–1558 (2018).
Gulati, M., Singh, S. K., Corrie, L., Kaur, I. P. & Chandwani, L. Delivery routes for faecal microbiota transplants: available, anticipated and aspired. Pharmacol. Res 159, 104954 (2020).
Ng, S. C. et al. Microbiota engraftment after faecal microbiota transplantation in obese subjects with type 2 diabetes: a 24-week, double-blind, randomised controlled trial. Gut 71, 716–723 (2022).
Ianiro, G. et al. Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat. Med. 28, 1913–1923 (2022).
Bajaj, J. S. et al. Fecal microbial transplant capsules are safe in hepatic encephalopathy: a phase 1, randomized, placebo-controlled trial. Hepatology 70, 1690–1703 (2019).
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Feuerstadt, P. et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 386, 220–229 (2022).
Hsu, C. L., Duan, Y., Fouts, D. E. & Schnabl, B. Intestinal virome and therapeutic potential of bacteriophages in liver disease. J. Hepatol. 75, 1465–1475 (2021).
Kurtz, C. B. et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl Med. 11, eaau7975 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03447730 (2021).
Russell, B. J. et al. Intestinal transgene delivery with native E. coli chassis allows persistent physiological changes. Cell 185, 3263–3277.e15 (2022).
Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667 (2021).
Yang, Z. et al. Inulin intervention attenuates hepatic steatosis in rats via modulating gut microbiota and maintaining intestinal barrier function. Food Res. Int. 163, 112309 (2023).
Huang, X. et al. Fructooligosaccharides attenuate non-alcoholic fatty liver disease by remodeling gut microbiota and association with lipid metabolism. Biomed. Pharmacother. 159, 114300 (2023).
Wrzosek, L. et al. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut 70, 1299–1308 (2021).
Pohl, K., Moodley, P. & Dhanda, A. The effect of increasing intestinal short-chain fatty acid concentration on gut permeability and liver injury in the context of liver disease: a systematic review. J. Gastroenterol. Hepatol. 37, 1498–1506 (2022).
Singh, V. et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell 175, 679–694.e22 (2018).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
Zhou, Y., Cui, Y. & Qu, X. Exopolysaccharides of lactic acid bacteria: structure, bioactivity and associations: a review. Carbohydr. Polym. 207, 317–332 (2019).
Pan, Z. et al. Postbiotics prepared using Lactobacillus paracasei CCFM1224 prevent nonalcoholic fatty liver disease by modulating the gut microbiota and liver metabolism. Int. J. Mol. Sci. 23, 13522 (2022).
Anstee, Q. M., Seth, D. & Day, C. P. Genetic factors that affect risk of alcoholic and nonalcoholic fatty liver disease. Gastroenterology 150, 1728–1744.e7 (2016).
Xue, L., Deng, Z., Luo, W., He, X. & Chen, Y. Effect of fecal microbiota transplantation on non-alcoholic fatty liver disease: a randomized clinical trial. Front. Cell Infect. Microbiol. 12, 759306 (2022).
Sharma, A. et al. Fecal microbiota transplantation in alcohol-associated acute-on-chronic liver failure: an open-label clinical trial. Hepatol. Int. 16, 433–446 (2022).
Craven, L. et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am. J. Gastroenterol. 115, 1055–1065 (2020).
Witjes, J. J. et al. Donor fecal microbiota transplantation alters gut microbiota and metabolites in obese individuals with steatohepatitis. Hepatol. Commun. 4, 1578–1590 (2020).
Bajaj, J. S. et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: a randomized clinical trial. Hepatology 66, 1727–1738 (2017). To our knowledge, this study is the first human randomized clinical trial using FMT for the treatment of hepatic encephalopathy in patients with cirrhosis and shows a reduction in hospitalizations due to liver-related complications.
Manzhalii, E. et al. Effect of a specific Escherichia coli Nissle 1917 strain on minimal/mild hepatic encephalopathy treatment. World J. Hepatol. 14, 634–646 (2022).
Zhu, W., Yan, M., Cao, H., Zhou, J. & Xu, Z. Effects of Clostridium butyricum capsules combined with rosuvastatin on intestinal flora, lipid metabolism, liver function and inflammation in NAFLD patients. Cell. Mol. Biol. 68, 64–69 (2022).
Mohamad Nor, M. H. et al. The effect of probiotics (MCP® BCMC® strains) on hepatic steatosis, small intestinal mucosal immune function, and intestinal barrier in patients with non-alcoholic fatty liver disease. Nutrients 13, 3192 (2021).
Chong, P. L., Laight, D., Aspinall, R. J., Higginson, A. & Cummings, M. H. A randomised placebo controlled trial of VSL#3® probiotic on biomarkers of cardiovascular risk and liver injury in non-alcoholic fatty liver disease. BMC Gastroenterol. 21, 144 (2021).
Behrouz, V., Aryaeian, N., Zahedi, M. J. & Jazayeri, S. Effects of probiotic and prebiotic supplementation on metabolic parameters, liver aminotransferases, and systemic inflammation in nonalcoholic fatty liver disease: a randomized clinical trial. J. Food Sci. 85, 3611–3617 (2020).
Scorletti, E. et al. Synbiotics alter fecal microbiomes, but not liver fat or fibrosis, in a randomized trial of patients with nonalcoholic fatty liver disease. Gastroenterology 158, 1597–1610.e7 (2020).
Ahn, S. B. et al. Randomized, double-blind, placebo-controlled study of a multispecies probiotic mixture in nonalcoholic fatty liver disease. Sci. Rep. 9, 5688 (2019).
Kobyliak, N. et al. A multi-strain probiotic reduces the fatty liver index, cytokines and aminotransferase levels in NAFLD patients: evidence from a randomized clinical trial. J. Gastrointestin. Liver Dis. 27, 41–49 (2018).
Manzhalii, E., Virchenko, O., Falalyeyeva, T., Beregova, T. & Stremmel, W. Treatment efficacy of a probiotic preparation for non-alcoholic steatohepatitis: a pilot trial. J. Dig. Dis. 18, 698–703 (2017).
Famouri, F., Shariat, Z., Hashemipour, M., Keikha, M. & Kelishadi, R. Effects of probiotics on nonalcoholic fatty liver disease in obese children and adolescents. J. Pediatr. Gastroenterol. Nutr. 64, 413–417 (2017).
Harrison, S. A. et al. A structurally optimized FXR agonist, MET409, reduced liver fat content over 12 weeks in patients with non-alcoholic steatohepatitis. J. Hepatol. 75, 25–33 (2021).
Harrison, S. A. et al. Efficacy and safety of aldafermin, an engineered FGF19 analog, in a randomized, double-blind, placebo-controlled trial of patients with nonalcoholic steatohepatitis. Gastroenterology 160, 219–231.e1 (2021). To our knowledge, this study is the first randomized clinical trial showing that an engineered FGF19 analogue could improve hepatic steatosis and liver injury in patients with NASH.
Newsome, P. N. et al. Volixibat in adults with non-alcoholic steatohepatitis: 24-week interim analysis from a randomized, phase II study. J. Hepatol. 73, 231–240 (2020).
Chambers, E. S. et al. The effects of dietary supplementation with inulin and inulin-propionate ester on hepatic steatosis in adults with non-alcoholic fatty liver disease. Diabetes Obes. Metab. 21, 372–376 (2019).
Traussnigg, S. et al. Norursodeoxycholic acid versus placebo in the treatment of non-alcoholic fatty liver disease: a double-blind, randomised, placebo-controlled, phase 2 dose-finding trial. Lancet Gastroenterol. Hepatol. 4, 781–793 (2019). To our knowledge, this study is the first randomized clinical trial showing that supplementation of a modified secondary bile acid in patients with NAFLD improved hepatic injury.
Harrison, S. A. et al. NGM282 for treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 391, 1174–1185 (2018).
Bernard, B. et al. Antibiotic prophylaxis for the prevention of bacterial infections in cirrhotic patients with gastrointestinal bleeding: a meta-analysis. Hepatology 29, 1655–1661 (1999).
Moon, A. M., Dominitz, J. A., Ioannou, G. N., Lowy, E. & Beste, L. A. Use of antibiotics among patients with cirrhosis and upper gastrointestinal bleeding is associated with reduced mortality. Clin. Gastroenterol. Hepatol. 14, 1629–1637.e1 (2016).
Gluud, L. L., Vilstrup, H. & Morgan, M. Y. Non-absorbable disaccharides versus placebo/no intervention and lactulose versus lactitol for the prevention and treatment of hepatic encephalopathy in people with cirrhosis. Cochrane Database Syst. Rev. 2016, CD003044 (2016).
Baxter, N. T. et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10, e02566-18 (2019).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Tang, W. H. & Hazen, S. L. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl. Res. 179, 108–115 (2017).
Witkowski, M., Weeks, T. L. & Hazen, S. L. Gut microbiota and cardiovascular disease. Circ. Res. 127, 553–570 (2020).
Acknowledgements
C.L.H. is supported by a Pilot and Feasibility Award from the Southern California Research Center for ALPD and Cirrhosis P50 AA011999 and by T32 DK007202. The Review article was supported by services provided by National Institutes of Health (NIH) centre P30 DK120515.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
B.S. has consulted for Ambys Medicines, Ferring Research Institute, Gelesis, HOST Therabiomics, Intercept Pharmaceuticals, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda, and is founder of Nterica Bio. UC San Diego has filed several patents with B.S. as inventor, and UC San Diego has received research support from Axial Biotherapeutics, BiomX, CymaBay Therapeutics, NGM Biopharmaceuticals, Prodigy Biotech and Synlogic Operating Company. C.L.H. declares no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hsu, C.L., Schnabl, B. The gut–liver axis and gut microbiota in health and liver disease. Nat Rev Microbiol 21, 719–733 (2023). https://doi.org/10.1038/s41579-023-00904-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-023-00904-3
This article is cited by
-
Single nuclear RNA sequencing of terminal ileum in patients with cirrhosis demonstrates multi-faceted alterations in the intestinal barrier
Cell & Bioscience (2024)
-
From NAFLD to MASLD: implications of the new nomenclature for preclinical and clinical research
Nature Metabolism (2024)
-
Application of Microbiome-Based Therapies in Chronic Respiratory Diseases
Journal of Microbiology (2024)
-
A new generation of mesenchymal stromal/stem cells differentially trained by immunoregulatory probiotics in a lupus microenvironment
Stem Cell Research & Therapy (2023)