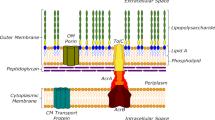
Abstract
Infections arising from multidrug-resistant pathogenic bacteria are spreading rapidly throughout the world and threaten to become untreatable. The origins of resistance are numerous and complex, but one underlying factor is the capacity of bacteria to rapidly export drugs through the intrinsic activity of efflux pumps. In this Review, we describe recent advances that have increased our understanding of the structures and molecular mechanisms of multidrug efflux pumps in bacteria. Clinical and laboratory data indicate that efflux pumps function not only in the drug extrusion process but also in virulence and the adaptive responses that contribute to antimicrobial resistance during infection. The emerging picture of the structure, function and regulation of efflux pumps suggests opportunities for countering their activities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
18 July 2018
In the version of this Review originally published, the author contributions of co-author Arthur Neuberger were incorrectly listed. The author contributions should have appeared as ‘D.D., X.W.-K., A.N., H.W.v.V., K.M.P., L.J.V.P. and B.F.L. researched data for the article, made substantial contributions to discussions of the content, wrote the article, and reviewed and edited the manuscript before submission’. This has now been corrected in all versions of the Review. The authors apologize to readers for this error.
References
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P. & Martínez, J. L. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 7, 1483 (2016).
Blair, J. M. et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl Acad. Sci. USA 112, 3511–3516 (2015).
Bergmiller, T. et al. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science 356, 311–315 (2017).
Li, X.-Z., Plésiat, P. & Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418 (2015).
Nichols, R. J. et al. Phenotypic landscape of a bacterial cell. Cell 144, 143–156 (2011).
Lee, A. et al. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 182, 3142–3150 (2000).
Tal, N. & Schuldiner, S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl Acad. Sci. USA 106, 9051–9056 (2009).
Hassan, K. A. et al. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl Acad. Sci. USA 110, 20254–20259 (2013).
Hassan, K. A., Liu, Q., Henderson, P. J. & Paulsen, I. T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6, e01982–14 (2015).
Fraimow, H. S. & Tsigrelis, C. Antimicrobial resistance in the intensive care unit: mechanisms, epidemiology, and management of specific resistant pathogens. Crit. Care Clin. 27, 163–205 (2011).
Viale, P., Giannella, M., Tedeschi, S. & Lewis, R. Treatment of MDR-Gram negative infections in the 21st century: a never ending threat for clinicians. Curr. Opin. Pharmacol. 24, 30–37 (2015).
Pu, Y. et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell 62, 284–294 (2016).
Li, N. et al. Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014).
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180 (2006).
Jin, M. S., Oldham, M. L., Zhang, Q. & Chen, J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566 (2012).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017).
Kodan, A. et al. Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc. Natl Acad. Sci. USA 111, 4049–4054 (2014).
Verhalen, B. et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017).
Hürlimann, L. M., Hohl, M. & Seeger, M. A. Split tasks of asymmetric nucleotide-binding sites in the heterodimeric ABC exporter EfrCD. FEBS J. 284, 1672–1687 (2017).
Mishra, S. et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 3, e02740 (2014).
Khare, D., Oldham, M. L., Orelle, C., Davidson, A. L. & Chen, J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell 33, 528–536 (2009).
Woo, J.-S., Zeltina, A., Goetz, B. A. & Locher, K. P. X-Ray structure of the Yersinia pestis heme transporter HmuUV. Nat. Struct. Mol. Biol. 19, 1310 (2012).
Korkhov, V. M., Mireku, S. A. & Locher, K. P. Structure of AMP-PNP-bound vitamin B sub 12 transporter BtuCD-F. Nature 490, 367 (2012).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Bountra, K. et al. Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 36, 3062–3079 (2017).
Venter, H., Shilling, R. A., Velamakanni, S., Balakrishnan, L. & van Veen, H. W. An ABC transporter with a secondary-active multidrug translocator domain. Nature 426, 866–870 (2003).
Singh, H. et al. ATP-dependent substrate transport by the ABC transporter MsbA is proton-coupled. Nat. Commun. 7, 12387 (2016).
van Veen, H. W. in ABC Transporters — 40 Years on (ed. George, A. M.) 37–51 (Springer, 2016).
Fitzpatrick, A. W. et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2, 17070 (2017).
Lin, H. T. et al. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J. Biol. Chem. 284, 1145–1154 (2009).
Okada, U. et al. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 8, 1336 (2017).
Crow, A., Greene, N. P., Kaplan, E. & Koronakis, V. Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl Acad. Sci. USA 114, 12572–12577 (2017).
Yamanaka, H., Kobayashi, H., Takahashi, E. & Okamoto, K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J. Bacteriol. 190, 7693–7698 (2008).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433 (2015).
Radestock, S. & Forrest, L. R. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 407, 698–715 (2011).
Kaback, H. R. A chemiosmotic mechanism of symport. Proc. Natl Acad. Sci. USA 112, 1259–1264 (2015).
Yin, Y., He, X., Szewczyk, P., Nguyen, T. & Chang, G. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744 (2006).
Heng, J. et al. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 25, 1060 (2015).
Jiang, D. et al. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc. Natl Acad. Sci. USA 110, 14664–14669 (2013).
Wisedchaisri, G., Park, M.-S., Iadanza, M. G., Zheng, H. & Gonen, T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 5, 4521 (2014).
Quistgaard, E. M., Löw, C., Moberg, P., Trésaugues, L. & Nordlund, P. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nat. Struct. Mol. Biol. 20, 766 (2013).
Zhao, Y. et al. Crystal structure of the E. coli peptide transporter YbgH. Structure 22, 1152–1160 (2014).
Fluman, N., Ryan, C. M., Whitelegge, J. P. & Bibi, E. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol. Cell 47, 777–787 (2012).
Masureel, M. et al. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat. Chem. Biol. 10, 149–155 (2014).
Norimatsu, Y., Hasegawa, K., Shimizu, N. & Toyoshima, C. Protein–phospholipid interplay revealed with crystals of a calcium pump. Nature 545, 193 (2017).
Ryan, R. M. & Vandenberg, R. J. Elevating the alternating-access model. Nat. Struct. Mol. Biol. 23, 187 (2016).
Martens, C. et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 23, 744 (2016).
Fluman, N., Adler, J., Rotenberg, S. A., Brown, M. H. & Bibi, E. Export of a single drug molecule in two transport cycles by a multidrug efflux pump. Nat. Commun. 5, 4615 (2014).
Schaedler, T. A. & van Veen, H. W. A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 24, 3653–3661 (2010).
Sun, J. et al. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73 (2014).
Federici, L. et al. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 Å resolution. J. Biol. Chem. 280, 15307–15314 (2005).
Mousa, J. J. et al. MATE transport of the E. coli-derived genotoxin colibactin. Nat. Microbiol. 1, 15009 (2016).
He, X. et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991 (2010).
Radchenko, M., Symersky, J., Nie, R. & Lu, M. Structural basis for the blockade of MATE multidrug efflux pumps. Nat. Commun. 6, 7995 (2015).
Tanaka, Y. et al. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247 (2013).
Ranaweera, I. et al. Structural comparison of bacterial multidrug efflux pumps of the major facilitator superfamily. Trends Cell. Mol. Biol. 10, 131 (2015).
Radchenko, M., Nie, R. & Lu, M. Disulfide cross-linking of a multidrug and toxic compound extrusion transporter impacts multidrug efflux. J. Biol. Chem. 291, 9818–9826 (2016).
Lu, M. et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl Acad. Sci. USA 110, 2099–2104 (2013).
Jin, Y., Nair, A. & van Veen, H. W. Multidrug transport protein NorM from Vibrio cholerae simultaneously couples to sodium-and proton-motive force. J. Biol. Chem. 289, 14624–14632 (2014).
Steed, P. R., Stein, R. A., Mishra, S., Goodman, M. C. & Mchaourab, H. S. Na+–substrate coupling in the multidrug antiporter NorM probed with a spin-labeled substrate. Biochemistry 52, 5790–5799 (2013).
Lu, M., Radchenko, M., Symersky, J., Nie, R. & Guo, Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20, 1310 (2013).
Kuk, A. C., Mashalidis, E. H. & Lee, S.-Y. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat. Struct. Mol. Biol. 24, 171 (2017).
Bolla, J. R. et al. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS ONE 9, e97903 (2014).
Eicher, T. et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl Acad. Sci. USA 109, 5687–5692 (2012).
Murakami, S., Nakashima, R., Yamashita, E. & Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593 (2002).
Nakashima, R., Sakurai, K., Yamasaki, S., Nishino, K. & Yamaguchi, A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569 (2011).
Oswald, C., Tam, H.-K. & Pos, K. M. Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat. Commun. 7, 13819 (2016).
Su, C.-C. et al. Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat. Commun. 8, 171 (2017).
Gong, X. et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 165, 1467–1478 (2016).
Tsukazaki, T. et al. Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235 (2011).
Kumar, N. et al. Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN. Proc. Natl Acad. Sci. USA 114, 6557–6562 (2017).
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T. & Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179 (2006).
Seeger, M. A., von Ballmoos, C., Verrey, F. & Pos, K. M. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48, 5801–5812 (2009).
Eicher, T. et al. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. elife 3, e03145 (2014).
Seeger, M. A. et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298 (2006).
Kim, H.-S. & Nikaido, H. Different functions of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB2C complex of Escherichia coli. Biochemistry 51, 4188–4197 (2012).
Sjuts, H. et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl Acad. Sci. USA 113, 3509–3514 (2016).
Wang, Z. et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife 6, e24905 (2017).
Nakashima, R. et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature 500, 102 (2013).
Hung, L.-W. et al. Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J. Struct. Funct. Genomics 14, 71–75 (2013).
Cha, H.-J., Müller, R. T. & Pos, K. M. Switch-loop flexibility affects transport of large drugs by the promiscuous AcrB multidrug efflux transporter. Antimicrob. Agents Chemother. 58, 4767–4772 (2014).
Yamaguchi, A., Nakashima, R. & Sakurai, K. Structural basis of RND-type multidrug exporters. Front. Microbiol. 6, 327 (2015).
Kinana, A. D., Vargiu, A. V. & Nikaido, H. Some ligands enhance the efflux of other ligands by the Escherichia coli multidrug pump AcrB. Biochemistry 52, 8342–8351 (2013).
Vargiu, A. V. et al. Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochim. Biophys. Acta 1862, 836–845 (2018).
Sennhauser, G., Amstutz, P., Briand, C., Storchenegger, O. & Grütter, M. G. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 5, e7 (2006).
Pos, K. M. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 1794, 782–793 (2009).
Ababou, A. New insights into the structural and functional involvement of the gate loop in AcrB export activity. Biochim. Biophys. Acta 1866, 242–253 (2018).
Zwama, M. et al. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 9, 124 (2018).
Ramaswamy, V. K., Vargiu, A. V., Malloci, G., Dreier, J. & Ruggerone, P. Molecular rationale behind the differential substrate specificity of bacterial RND multi-drug transporters. Sci. Rep. 7, 8075 (2017).
Schumacher, M. A., Miller, M. C. & Brennan, R. G. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 23, 2923–2930 (2004).
Nikaido, H., Basina, M., Nguyen, V. & Rosenberg, E. Y. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180, 4686–4692 (1998).
Neyfakh, A. A. Mystery of multidrug transporters: the answer can be simple. Mol. Microbiol. 44, 1123–1130 (2002).
Bay, D. C., Rommens, K. L. & Turner, R. J. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778, 1814–1838 (2008).
Fleishman, S. J. et al. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 364, 54–67 (2006).
Chen, Y.-J. et al. X-ray structure of EmrE supports dual topology model. Proc. Natl Acad. Sci. USA 104, 18999–19004 (2007).
Morrison, E. A. et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45 (2012).
Fluman, N., Tobiasson, V. & von Heijne, G. Stable membrane orientations of small dual-topology membrane proteins. Proc. Natl Acad. Sci. USA 114, 7987–7992 (2017).
Woodall, N. B., Yin, Y. & Bowie, J. U. Dual-topology insertion of a dual-topology membrane protein. Nat. Commun. 6, 8099 (2015).
Dastvan, R., Fischer, A. W., Mishra, S., Meiler, J. & Mchaourab, H. S. Protonation-dependent conformational dynamics of the multidrug transporter EmrE. Proc. Natl Acad. Sci. USA 113, 1220–1225 (2016).
Gayen, A., Leninger, M. & Traaseth, N. J. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat. Chem. Biol. 12, 141 (2016).
Lytvynenko, I., Brill, S., Oswald, C. & Pos, K. M. Molecular basis of polyspecificity of the small multidrug resistance efflux pump AbeS from Acinetobacter baumannii. J. Mol. Biol. 428, 644–657 (2016).
Brill, S., Sade-Falk, O., Elbaz-Alon, Y. & Schuldiner, S. Specificity determinants in small multidrug transporters. J. Mol. Biol. 427, 468–477 (2015).
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O. & Piddock, L. J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42 (2015).
Koteva, K. et al. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6, 327–329 (2010).
Fritz, G. et al. A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6, e00975 (2015).
Piepenbreier, H., Fritz, G. & Gebhard, S. Transporters as information processors in bacterial signalling pathways. Mol. Microbiol. 104, 1–15 (2017).
Gushchin, I. et al. Mechanism of transmembrane signaling by sensor histidine kinases. Science 356, eaah6345 (2017).
Zschiedrich, C. P., Keidel, V. & Szurmant, H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 (2016).
Poole, K. et al. Potentiation of aminoglycoside activity in Pseudomonas aeruginosa by targeting the AmgRS envelope stress-responsive two-component system. Antimicrob. Agents Chemother. 60, 3509–3518 (2016).
Sun, J.-R. et al. Single amino acid substitution Gly186Val in AdeS restores tigecycline susceptibility of Acinetobacter baumannii. J. Antimicrob. Chemother. 71, 1488–1492 (2016).
Marchand, I., Damier-Piolle, L., Courvalin, P. & Lambert, T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48, 3298–3304 (2004).
Chang, T.-Y. et al. AdeR protein regulates adeABC expression by binding to a direct-repeat motif in the intercistronic spacer. Microbiol. Res. 183, 60–67 (2016).
Nowak, J., Schneiders, T., Seifert, H. & Higgins, P. G. The Asp20-to-Asn substitution in the response regulator AdeR leads to enhanced efflux activity of AdeB in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 1085–1090 (2016).
Richmond, G. E. et al. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7, e00430–16 (2016).
Nishino, K., Nikaido, E. & Yamaguchi, A. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189, 9066–9075 (2007).
Nishino, K. & Yamaguchi, A. Overexpression of the response RegulatorevgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183, 1455–1458 (2001).
Casino, P., Rubio, V. & Marina, A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009).
Chen, H. et al. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl Acad. Sci. USA 105, 13586–13591 (2008).
Li, M. et al. Crystal structure of the transcriptional regulator AcrR from Escherichia coli. J. Mol. Biol. 374, 591–603 (2007).
Wilke, M. S. et al. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc. Natl Acad. Sci. USA 105, 14832–14837 (2008).
Yamada, J. et al. Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli. J. Antimicrob. Chemother. 65, 853–858 (2010).
Vidyaprakash, E., Abrams, A. J., Shafer, W. M. & Trees, D. L. Whole genome sequencing of a large panel of contemporary Neisseria gonorrhoeae clinical isolates indicates that a wild-type mtrA gene is common: implications for inducible antimicrobial resistance. Antimicrob. Agents Chemother. 61, e00262–17 (2017).
Sharma, P. et al. The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat. Commun. 8, 1444 (2017).
Dersch, P., Khan, M. A., Mühlen, S. & Görke, B. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803 (2017).
Lalaouna, D., Eyraud, A., Chabelskaya, S., Felden, B. & Masse, E. Regulatory RNAs involved in bacterial antibiotic resistance. PLoS Pathog. 10, e1004299 (2014).
Dar, D. & Sorek, R. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Curr. Opin. Microbiol. 36, 111–117 (2017).
Parker, A. & Gottesman, S. Small RNA regulation of TolC, the outer membrane component of bacterial multidrug transporters. J. Bacteriol. 198, 1101–1113 (2016).
Nishino, K., Yamasaki, S., Hayashi-Nishino, M. & Yamaguchi, A. Effect of overexpression of small non-coding DsrA RNA on multidrug efflux in Escherichia coli. J. Antimicrob. Chemother. 66, 291–296 (2010).
Jackson, L. A., Pan, J.-C., Day, M. W. & Dyer, D. W. Control of RNA stability by NrrF, an iron-regulated small RNA in Neisseria gonorrhoeae. J. Bacteriol. 195, 5166–5173 (2013).
Göpel, Y. & Görke, B. Rewiring two-component signal transduction with small RNAs. Curr. Opin. Microbiol. 15, 132–139 (2012).
Storz, G., Wolf, Y. I. & Ramamurthi, K. S. Small proteins can no longer be ignored. Annu. Rev. Biochem. 83, 753–777 (2014).
Hobbs, E. C., Yin, X., Paul, B. J., Astarita, J. L. & Storz, G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl Acad. Sci. USA 109, 16696–16701 (2012).
Du, D. et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515 (2014).
Young, B. C. et al. Severe infections emerge from commensal bacteria by adaptive evolution. eLife 6, e30637 (2017).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K.(p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013).
Yang, S. et al. Antibiotic regimen based on population analysis of residing persister cells eradicates Staphylococcus epidermidis biofilms. Sci. Rep. 5, 18578 (2015).
Everett, M. J., Jin, Y. F., Ricci, V. & Piddock, L. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40, 2380–2386 (1996).
Kao, C.-Y. et al. Molecular characterization of antimicrobial susceptibility of Salmonella isolates: first identification of a plasmid carrying qnrD or oqxAB in Taiwan. J. Microbiol. Immunol. Infect. 50, 214–223 (2017).
Machado, D. et al. Interplay between mutations and efflux in drug resistant clinical isolates of Mycobacterium tuberculosis. Front. Microbiol. 8, 711 (2017).
Zampieri, M. et al. Metabolic constraints on the evolution of antibiotic resistance. Mol. Syst. Biol. 13, 917 (2017).
Ohneck, E. A. et al. Overproduction of the MtrCDE efflux pump in Neisseria gonorrhoeae produces unexpected changes in cellular transcription patterns. Antimicrob. Agents Chemother. 59, 724–726 (2015).
Pacheco, J. O., Alvarez-Ortega, C., Rico, M. A. & Martínez, J. L. Metabolic compensation of fitness costs is a general outcome for antibiotic-resistant Pseudomonas aeruginosa mutants overexpressing efflux pumps. mBio 8, e00500–17 (2017).
Stickland, H. G., Davenport, P. W., Lilley, K. S., Griffin, J. L. & Welch, M. Mutation of nfxB causes global changes in the physiology and metabolism of Pseudomonas aeruginosa. J. Proteome Res. 9, 2957–2967 (2010).
Wang-Kan, X. et al. Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar typhimurium. mBio 8, e00968–17 (2017).
Bailey, A. M. et al. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192, 1607–1616 (2010).
De Majumdar, S. et al. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog. 11, e1004627 (2015).
Yao, H. et al. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7, e01543–16 (2016).
González-Pasayo, R. & Martínez-Romero, E. Multiresistance genes of Rhizobium etli CFN42. Mol. Plant. Microbe Interact. 13, 572–577 (2000).
Thanassi, D. G., Cheng, L. W. & Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518 (1997).
Elbourne, L. D., Tetu, S. G., Hassan, K. A. & Paulsen, I. T. TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324 (2016).
Lee, E. H. & Shafer, W. M. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33, 839–845 (1999).
Nishino, K., Latifi, T. & Groisman, E. A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141 (2006).
Buckley, A. M. et al. The AcrAB–TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8, 847–856 (2006).
Bogomolnaya, L. M. et al. The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. mBio 4, e00630–13 (2013).
Kunkle, D. E., Bina, X. R. & Bina, J. E. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8, e00126–17 (2017).
Horiyama, T. & Nishino, K. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS ONE 9, e108642 (2014).
Sachla, A. J. & Eichenbaum, Z. The GAS PefCD exporter is a MDR system that confers resistance to heme and structurally diverse compounds. BMC Microbiol. 16, 68 (2016).
Hagman, K. E. et al. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141, 611–622 (1995).
Kobayashi, N., Nishino, K. & Yamaguchi, A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644 (2001).
Adams, K. N. et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53 (2011).
Lee, J. & Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41 (2015).
Minagawa, S. et al. RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. 12, 70 (2012).
Lamarche, M. G. & Déziel, E. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS ONE 6, e24310 (2011).
Moore, J. D., Gerdt, J. P., Eibergen, N. R. & Blackwell, H. E. Active efflux influences the potency of quorum sensing inhibitors in Pseudomonas aeruginosa. Chembiochem 15, 435–442 (2014).
Sakhtah, H. et al. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Natl Acad. Sci. USA 113, E3538–E3547 (2016).
Aoki, S. K. et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70, 323–340 (2008).
Ruhe, Z. C., Wallace, A. B., Low, D. A. & Hayes, C. S. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio 4, e00480–13 (2013).
Yoshida, T., Qin, L., Egger, L. A. & Inouye, M. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281, 17114–17123 (2006).
Dar, D. et al. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352, aad9822 (2016).
Lin, M.-F., Lin, Y.-Y., Tu, C.-C. & Lan, C.-Y. Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infection 50, 224–231 (2017).
Srinivasan, V. B., Rajamohan, G. & Gebreyes, W. A. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 5312–5316 (2009).
Podnecky, N. L., Wuthiekanun, V., Peacock, S. J. & Schweizer, H. P. The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob. Agents Chemother. 57, 4381–4386 (2013).
Swick, M. C., Morgan-Linnell, S. K., Carlson, K. M. & Zechiedrich, L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob. Agents Chemother. 55, 921–924 (2011).
Hansen, L. H., Jensen, L. B., Sørensen, H. I. & Sørensen, S. J. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60, 145–147 (2007).
Doran, J. L. et al. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab. Immunol. 4, 23–32 (1997).
Andries, K. et al. Acquired resistancxe of Mycobacterium tuberculosis to bedaquiline. PLoS ONE 9, e102135 (2014).
Rodrigues, L., Villellas, C., Bailo, R., Viveiros, M. & Aínsa, J. A. Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751–757 (2013).
Dreier, J. & Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 6, 660 (2015).
Li, X.-Z., Poole, K. & Nikaido, H. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 47, 27–33 (2003).
Golparian, D., Shafer, W. M., Ohnishi, M. & Unemo, M. Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 58, 3556–3559 (2014).
Schindler, B. D. & Kaatz, G. W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updat. 27, 1–13 (2016).
Acknowledgements
The authors thank S. Murakami, B. Görke, J. Blaza, M. Welch, A. Vargiu, P. Ruggerone, L. Schmitt, and M. Osman for helpful discussions and the reviewers for helpful comments. B.F.L. and D.D. are supported by the Wellcome Trust and the European Research Council (742210). K.M.P. is supported by the German Research Foundation (DFG-SFB 807, ‘Transport and Communication across Biological Membranes’, and DFG-FOR2251, ‘Adaptation and Persistence of the Emerging Pathogen Acinetobacter baumannii), the DFG-EXC115 (Cluster of Excellence Frankfurt—Macromolecular Complexes), the Innovative Medicines Joint Undertaking (IMI-Translocation) under grant agreement no. 115525 and the National Institute of Allergy and Infectious Diseases (grant R44 AI100332). H.W.v.V., K.M.P. and B.F.L. are supported by a grant from the Human Frontier Science Program (RGP0034/2013). H.W.v.V. is also supported by the Biotechnology and Biological Sciences Research Council (grant BB/R00224X/1). A.N. is a recipient of a Herchel Smith Scholarship. X.W.-K. is supported by Consejo Nacional de Ciencia y Tecnología (CONACyT), Mexico. L.J.V.P. is supported by the Biotechnology and Biological Sciences Research Council (grant BB/N014200/1) and the Medical Research Council (MR/022596/1).
Reviewer information
Nature Reviews Microbiology thanks K. Beis, H. Schweizer and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
D.D., X.W.-K., A.N., H.W.v.V., K.M.P., L.J.V.P. and B.F.L. researched data for the article, made substantial contributions to discussions of the content, wrote the article, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Transporter classification database: http://www.tcdb.org
Supplementary information
Rights and permissions
About this article
Cite this article
Du, D., Wang-Kan, X., Neuberger, A. et al. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16, 523–539 (2018). https://doi.org/10.1038/s41579-018-0048-6
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0048-6
This article is cited by
-
Non-antibiotic pharmaceuticals are toxic against Escherichia coli with no evolution of cross-resistance to antibiotics
npj Antimicrobials and Resistance (2024)
-
Multidrug efflux in Gram-negative bacteria: structural modifications in active compounds leading to efflux pump avoidance
npj Antimicrobials and Resistance (2024)
-
Discovery and description of novel phage genomes from urban microbiomes sampled by the MetaSUB consortium
Scientific Reports (2024)
-
High-throughput transcriptomics of 409 bacteria–drug pairs reveals drivers of gut microbiota perturbation
Nature Microbiology (2024)
-
Modulators of Candida albicans Membrane Drug Transporters: A Lucrative Portfolio for the Development of Effective Antifungals
Molecular Biotechnology (2024)