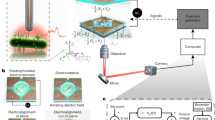
Abstract
Biointerfaces mediate safe and efficient cell manipulation, which is essential for biomedical innovations in advanced therapies and diagnostics. The biointerface established by vertical nanoprobes — arrays of vertical high-aspect-ratio nanostructures — has emerged as a simple, controllable and powerful tool for interrogating and manipulating cells. Vertical nanoprobes have substantially improved our ability to control and characterize the intracellular environment, guide biophysical stimuli with nanoscale precision to defined cell compartments, stimulate and record the electrical activity of cells, and transport hard-to-deliver drugs. These capabilities are enabling substantial advances in bioelectronics, spatiotemporally resolved molecular diagnostics, and cell and gene therapy — all underpinned by the design versatility of the nanoprobe biointerface. This Review discusses how the design of a vertical nanoprobe biointerface determines its ability to interrogate and control a cell.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lane, S. W., Williams, D. A. & Watt, F. M. Modulating the stem cell niche for tissue regeneration. Nat. Biotechnol. 32, 795–803 (2014).
Wang, N., Tytell, J. D. & Ingber, D. E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009).
Hulshof, F. F. et al. NanoTopoChip: high-throughput nanotopographical cell instruction. Acta Biomater. 62, 188–198 (2017).
Patel, N. et al. Spatially controlled cell engineering on biodegradable polymer surfaces. FASEB J. 12, 1447–1454 (1998).
Downing, T. L. et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 12, 1154–1162 (2013).
Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016).
Yu, Y. & Yoshimura, S. H. Investigating the morphological dynamics of the plasma membrane by high-speed atomic force microscopy. J. Cell Sci. 134, jcs243584 (2021).
Paulitschke, P. et al. Ultraflexible nanowire array for label- and distortion-free cellular force tracking. Nano Lett. 19, 2207–2214 (2019).
O’Brien, J. A. & Lummis, S. C. Biolistic transfection of neuronal cultures using a hand-held gene gun. Nat. Protoc. 1, 977 (2006).
McMurray, R. J. et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat. Mater. 10, 637–644 (2011).
Venslauskas, M. S. & Šatkauskas, S. Mechanisms of transfer of bioactive molecules through the cell membrane by electroporation. Eur. Biophys. J. 44, 277–289 (2015).
Nath, A. R., Chen, R. H. & Stanley, E. F. Cryoloading: introducing large molecules into live synaptosomes. Front. Cell. Neurosci. 8, 4 (2014).
Antkowiak, M. et al. Fast targeted gene transfection and optogenetic modification of single neurons using femtosecond laser irradiation. Sci. Rep. 3, 3281 (2013).
Ramesan, S. et al. Acoustically-mediated intracellular delivery. Nanoscale 10, 13165–13178 (2018).
Lichtenberg, D., Ahyayauch, H. & Goñi, F. M. The mechanism of detergent solubilization of lipid bilayers. Biophys. J. 105, 289–299 (2013).
Korzh, V. & Strähle, U. Marshall Barber and the century of microinjection: from cloning of bacteria to cloning of everything. Differentiation 70, 221–226 (2002).
Sakmann, B. & Neher, E. Patch clamp techniques for studying ionic channels in excitable membranes. Annu. Rev. Physiol. 46, 455–472 (1984).
Lee, S. E. et al. Remote optical switch for localized and selective control of gene interference. Nano Lett. 9, 562–570 (2009).
Shin, H. et al. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat. Commun. 12, 492 (2021).
Cai, P. et al. Combinatorial nano–bio interfaces. ACS Nano 12, 5078–5084 (2018).
Nadeem, D. et al. Embossing of micropatterned ceramics and their cellular response. J. Biomed. Mater. Res. Part A 101, 3247–3255 (2013).
Hondrich, T. J. et al. MEA recordings and cell–substrate investigations with plasmonic and transparent, tunable holey gold. ACS Appl. Mater. Interfaces 11, 46451–46461 (2019).
Elnathan, R. et al. The start-ups taking nanoneedles into the clinic. Nat. Nanotechnol. https://doi.org/10.1038/s41565-022-01158-5 (2022).
Li, X. et al. Vertical nanowire array-based biosensors: device design strategies and biomedical applications. J. Mater. Chem. B 8, 7609–7632 (2020).
Lin, Z. C. et al. Iridium oxide nanotube electrodes for sensitive and prolonged intracellular measurement of action potentials. Nat. Commun. 5, 3206 (2014).
Shiu, J.-Y. et al. Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat. Cell Biol. 20, 262–271 (2018).
Cao, Y. et al. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl Acad. Sci. USA 114, E1866–E1874 (2017).
Chiappini, C. et al. Biodegradable nanoneedles for localized delivery of nanoparticles in vivo: exploring the biointerface. ACS Nano 9, 5500–5509 (2015).
Hansel, C. S. et al. Nanoneedle-mediated stimulation of cell mechanotransduction machinery. ACS Nano 13, 2913–2926 (2019).
Chiappini, C. et al. Mapping local cytosolic enzymatic activity in human esophageal mucosa with porous silicon nanoneedles. Adv. Mater. 27, 5147–5152 (2015).
Na, Y.-R. et al. Probing enzymatic activity inside living cells using a nanowire–cell ‘sandwich’ assay. Nano Lett. 13, 153–158 (2013).
Krivitsky, V. et al. Si nanowires forest-based on-chip biomolecular filtering, separation and preconcentration devices: nanowires do it all. Nano Lett. 12, 4748–4756 (2012).
Zhao, Y. et al. Scalable ultrasmall three-dimensional nanowire transistor probes for intracellular recording. Nat. Nanotechnol. 14, 783–790 (2019).
Robinson, J. T. et al. Vertical nanowire electrode arrays as a scalable platform for intracellular interfacing to neuronal circuits. Nat. Nanotechnol. 7, 180–184 (2012).
Chiappini, C. et al. Tutorial: using nanoneedles for intracellular delivery. Nat. Protoc. 16, 4539–4563 (2021).
Xie, X. et al. Nanostraw–electroporation system for highly efficient intracellular delivery and transfection. ACS Nano 7, 4351–4358 (2013).
Chiappini, C. et al. Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization. Nat. Mater. 14, 532–539 (2015).
Lestrell, E. et al. Engineered nano-bio interfaces for intracellular delivery and sampling: applications, agency and artefacts. Mater. Today 33, 87–104 (2020).
Chen, Y. et al. Emerging roles of 1D vertical nanostructures in orchestrating immune cell functions. Adv. Mater. 32, 2001668 (2020).
Buch-Månson, N. et al. Mapping cell behavior across a wide range of vertical silicon nanocolumn densities. Nanoscale 9, 5517–5527 (2017).
Kim, W. et al. Interfacing silicon nanowires with mammalian cells. J. Am. Chem. Soc. 129, 7228–7229 (2007).
Buch-Månson, N. et al. Towards a better prediction of cell settling on nanostructure arrays — simple means to complicated ends. Adv. Funct. Mater. 25, 3246–3255 (2015).
Xie, X. et al. Mechanical model of vertical nanowire cell penetration. Nano Lett. 13, 6002–6008 (2013).
Xie, X. et al. Determining the time window for dynamic nanowire cell penetration processes. Acs Nano 9, 11667–11677 (2015).
Li, C. et al. Large-scale, robust mushroom-shaped nanochannel array membrane for ultrahigh osmotic energy conversion. Sci. Adv. 7, eabg2183 (2021).
Wen, R. et al. Intracellular delivery and sensing system based on electroplated conductive nanostraw arrays. ACS Appl. Mater. Interfaces 11, 43936–43948 (2019).
Persson, H. et al. Fibroblasts cultured on nanowires exhibit low motility, impaired cell division, and DNA damage. Small 9, 4006–4016 (2013).
Persson, H. et al. From immobilized cells to motile cells on a bed-of-nails: effects of vertical nanowire array density on cell behaviour. Sci. Rep. 5, 18535 (2015).
Matsumoto, D. et al. Oscillating high-aspect-ratio monolithic silicon nanoneedle array enables efficient delivery of functional bio-macromolecules into living cells. Sci. Rep. 5, 15325 (2015).
Obataya, I. et al. Nanoscale operation of a living cell using an atomic force microscope with a nanoneedle. Nano Lett. 5, 27–30 (2005).
Kim, H. et al. Flexible elastomer patch with vertical silicon nanoneedles for intracellular and intratissue nanoinjection of biomolecules. Sci. Adv. 4, eaau6972 (2018).
Buch-Månson, N. et al. Rapid prototyping of polymeric nanopillars by 3D direct laser writing for controlling cell behavior. Sci. Rep. 7, 15325 (2017).
Yoh, H. Z. et al. Polymeric nanoneedle arrays mediate stiffness-independent intracellular delivery. Adv. Funct. Mater. 32, 2104828 (2022).
He, G. et al. Multifunctional branched nanostraw-electroporation platform for intracellular regulation and monitoring of circulating tumor cells. Nano Lett. 19, 7201–7209 (2019).
Elnathan, R. et al. Optically transparent vertical silicon nanowire arrays for live-cell imaging. J. Nanobiotechnol. 19, 51 (2021).
Kim, S. et al. 3D super-resolved imaging in live cells using sub-diffractive plasmonic localization of hybrid nanopillar arrays. Nanophotonics 9, 2847–2859 (2020).
Frederiksen, R. S. et al. Nanowire-aperture probe: local enhanced fluorescence detection for the investigation of live cells at the nanoscale. ACS Photonics 3, 1208–1216 (2016).
Wang, Y. et al. Poking cells for efficient vector-free intracellular delivery. Nat. Commun. 5, 4466 (2014).
Dipalo, M. et al. Cells adhering to 3D vertical nanostructures: cell membrane reshaping without stable internalization. Nano Lett. 18, 6100–6105 (2018).
Amin, H. et al. Biofunctionalized 3D nanopillar arrays fostering cell guidance and promoting synapse stability and neuronal activity in networks. ACS Appl. Mater. Interfaces 10, 15207–15215 (2018).
Santoro, F. et al. On chip guidance and recording of cardiomyocytes with 3D mushroom-shaped electrodes. Nano Lett. 13, 5379–5384 (2013).
Hai, A. et al. Spine-shaped gold protrusions improve the adherence and electrical coupling of neurons with the surface of micro-electronic devices. J. R. Soc. Interface 6, 1153–1165 (2009).
Wang, H. et al. in Non-Viral Gene Delivery Vectors (ed. Candiani, G.) 279–287 (Springer, 2016).
Sridar, S. et al. Peptide modification of polyimide-insulated microwires: towards improved biocompatibility through reduced glial scarring. Acta Biomater. 60, 154–166 (2017).
Duan, X. et al. Intracellular recordings of action potentials by an extracellular nanoscale field-effect transistor. Nat. Nanotechnol. 7, 174–179 (2012).
Almquist, B. D. & Melosh, N. A. Fusion of biomimetic stealth probes into lipid bilayer cores. Proc. Natl Acad. Sci. USA 107, 5815–5820 (2010).
Almquist, B. D. & Melosh, N. A. Molecular structure influences the stability of membrane penetrating biointerfaces. Nano Lett. 11, 2066–2070 (2011).
Vutti, S. et al. Click chemistry mediated functionalization of vertical nanowires for biological applications. Chem. Eur. J. 22, 496–500 (2016).
Kihara, T. et al. Nanoneedle surface modification with 2-methacryloyloxyethyl phosphorylcholine polymer to reduce nonspecific protein adsorption in a living cell. NanoBiotechnology 3, 127–134 (2007).
Qu, Y. et al. A universal platform for high-efficiency ‘engineering’ living cells: integration of cell capture, intracellular delivery of biomolecules, and cell harvesting functions. Adv. Funct. Mater. 30, 1906362 (2020).
Sahoo, P. K. et al. Nanowire arrays as cell force sensors to investigate adhesin-enhanced holdfast of single cell bacteria and biofilm stability. Nano Lett. 16, 4656–4664 (2016).
Leriche, G., Chisholm, L. & Wagner, A. Cleavable linkers in chemical biology. Bioorg. Med. Chem. 20, 571–582 (2012).
Shalek, A. K. et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl Acad. Sci. USA 107, 1870–1875 (2010).
Elnathan, R. et al. Maximizing transfection efficiency of vertically aligned silicon nanowire arrays. Adv. Funct. Mater. 25, 7215–7225 (2015).
Higgins, S. G. et al. High-aspect-ratio nanostructured surfaces as biological metamaterials. Adv. Mater. 32, 1903862 (2020).
He, G. et al. Nanoneedle platforms: the many ways to pierce the cell membrane. Adv. Funct. Mater. 30, 1909890 (2020).
Kawamura, R. et al. A new cell separation method based on antibody-immobilized nanoneedle arrays for the detection of intracellular markers. Nano Lett. 17, 7117–7124 (2017).
Fang, J. et al. Accurate and efficient intracellular delivery biosensing system by nanostrawed electroporation array. Biosens. Bioelectron. 194, 113583 (2021).
Messina, G. C. et al. Spatially, temporally, and quantitatively controlled delivery of broad range of molecules into selected cells through plasmonic nanotubes. Adv. Mater. 27, 7145–7149 (2015).
Dipalo, M. et al. 3D plasmonic nanoantennas integrated with MEA biosensors. Nanoscale 7, 3703–3711 (2015).
Zhao, W. et al. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat. Nanotechnol. 12, 750–756 (2017).
Hanson, L. et al. Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat. Nanotechnol. 10, 554–562 (2015).
Wei, Y. et al. Directing stem cell differentiation via electrochemical reversible switching between nanotubes and nanotips of polypyrrole array. ACS Nano 11, 5915–5924 (2017).
Beckwith, K. S. et al. Influence of nanopillar arrays on fibroblast motility, adhesion, and migration mechanisms. Small 15, 1902514 (2019).
Dai, J. et al. Cellular architecture response to aspect ratio tunable nanoarrays. Nanoscale 12, 12395–12404 (2020).
Park, J. et al. Nanosize and vitality: TiO2 nanotube diameter directs cell fate. Nano Lett. 7, 1686–1691 (2007).
VanDersarl, J. J., Xu, A. M. & Melosh, N. A. Nanostraws for direct fluidic intracellular access. Nano Lett. 12, 3881–3886 (2012).
Hanson, L. et al. Characterization of the cell–nanopillar interface by transmission electron microscopy. Nano Lett. 12, 5815–5820 (2012).
Santoro, F. et al. Revealing the cell–material interface with nanometer resolution by focused ion beam/scanning electron microscopy. ACS Nano 11, 8320–8328 (2017).
von Erlach, T. C. et al. Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nat. Mater. 17, 237–242 (2018).
Berthing, T. et al. Cell membrane conformation at vertical nanowire array interface revealed by fluorescence imaging. Nanotechnology 23, 415102 (2012).
Wang, Z. et al. Interrogation of cellular innate immunity by diamond-nanoneedle-assisted intracellular molecular fishing. Nano Lett. 15, 7058–7063 (2015).
He, G. et al. Hollow nanoneedle-electroporation system to extract intracellular protein repetitively and nondestructively. ACS Sens. 3, 1675–1682 (2018).
Dipalo, M. et al. Membrane poration mechanisms at the cell–nanostructure interface. Adv. Biosyst. 3, 1900148 (2019).
Arnold, M. et al. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem 5, 383–388 (2004).
Li, S. et al. Effects of nanoscale spatial arrangement of arginine–glycine–aspartate peptides on dedifferentiation of chondrocytes. Nano Lett. 15, 7755–7765 (2015).
Maheshwari, G. et al. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 113, 1677–1686 (2000).
Sun, Z., Guo, S. S. & Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016).
Yurugi, H. et al. A subset of flavaglines inhibits KRAS nanoclustering and activation. J. Cell Sci. 133, jcs244111 (2020).
Liang, H. et al. Membrane curvature sensing of the lipid-anchored K-Ras small GTPase. Life Sci. Alliance 2, e201900343 (2019).
Mu, H. W. et al. Patterning of oncogenic Ras clustering in live cells using vertically aligned nanostructure arrays. Nano Lett. 22, 1007–1016 (2022).
Galic, M. et al. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. eLife 3, e03116 (2014).
McMahon, H. T. & Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011).
Gopal, S. et al. Porous silicon nanoneedles modulate endocytosis to deliver biological payloads. Adv. Mater. 31, 1806788 (2019).
Aslanoglou, S. et al. Efficient transmission electron microscopy characterization of cell–nanostructure interfacial interactions. J. Am. Chem. Soc. 142, 15649–15653 (2020).
Chen, Y. et al. Silicon-nanotube-mediated intracellular delivery enables ex vivo gene editing. Adv. Mater. 32, 2000036 (2020).
Chen, Y. et al. Cellular deformations induced by conical silicon nanowire arrays facilitate gene delivery. Small 15, 1904819 (2019).
Peter, B. J. et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 (2004).
Galic, M. et al. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat. Cell Biol. 14, 874–881 (2012).
Su, M. et al. Comparative study of curvature sensing mediated by F-BAR and an intrinsically disordered region of FBP17. iScience 23, 101712 (2020).
Lou, H.-Y. et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc. Natl Acad. Sci. USA 116, 23143–23151 (2019).
Li, L.-L. et al. Nanobar array assay revealed complementary roles of BIN1 splice isoforms in cardiac T-tubule morphogenesis. Nano Lett. 20, 6387–6395 (2020).
Li, X. et al. Nanoscale surface topography reduces focal adhesions and cell stiffness by enhancing integrin endocytosis. Nano Lett. 21, 8518–8526 (2021).
Beckwith, K. S. et al. Tunable high aspect ratio polymer nanostructures for cell interfaces. Nanoscale 7, 8438–8450 (2015).
De Martino, S. et al. Dynamic manipulation of cell membrane curvature by light-driven reshaping of azopolymer. Nano Lett. 20, 577–584 (2019).
Qi, S. et al. Cell adhesion and spreading behavior on vertically aligned silicon nanowire arrays. ACS Appl. Mater. Interfaces 1, 30–34 (2009).
Liu, H. et al. TiO2 nanorod arrays with mesoscopic micro–nano interfaces for in situ regulation of cell morphology and nucleus deformation. ACS Appl. Mater. Interfaces 10, 66–74 (2018).
Aalipour, A. et al. Plasma membrane and actin cytoskeleton as synergistic barriers to nanowire cell penetration. Langmuir 30, 12362–12367 (2014).
Seong, H. et al. Size-tunable nanoneedle arrays for influencing stem cell morphology, gene expression, and nuclear membrane curvature. ACS Nano 14, 5371–5381 (2020).
Carthew, J. et al. Precision surface microtopography regulates cell fate via changes to actomyosin contractility and nuclear architecture. Adv. Sci. 8, 2003186 (2021).
Seo, J. et al. Neuro-taxis: neuronal movement in gradients of chemical and physical environments. Dev. Neurobiol. 80, 361–377 (2020).
Dong, Y. et al. Nanotechnology shaping stem cell therapy: recent advances, application, challenges, and future outlook. Biomed. Pharmacother. 137, 111236 (2021).
Yang, J. et al. Nanotopographical induction of osteogenesis through adhesion, bone morphogenic protein cosignaling, and regulation of microRNAs. ACS Nano 8, 9941–9953 (2014).
Qiu, J. et al. TiO2 nanorod array constructed nanotopography for regulation of mesenchymal stem cells fate and the realization of location-committed stem cell differentiation. Small 12, 1770–1778 (2016).
Kim, H. et al. Neuron-like differentiation of mesenchymal stem cells on silicon nanowires. Nanoscale 7, 17131–17138 (2015).
Lin, H.-I. et al. SiNWs biophysically regulate the fates of human mesenchymal stem cells. Sci. Rep. 8, 12913 (2018).
Kuo, S.-W. et al. Regulation of the fate of human mesenchymal stem cells by mechanical and stereo-topographical cues provided by silicon nanowires. Biomaterials 33, 5013–5022 (2012).
Kim, Y. J. et al. Association between cell microenvironment altered by gold nanowire array and regulation of partial epithelial-mesenchymal transition. Adv. Funct. Mater. 31, 2008758 (2021).
Rasmussen, C. H. et al. Enhanced differentiation of human embryonic stem cells toward definitive endoderm on ultrahigh aspect ratio nanopillars. Adv. Funct. Mater. 26, 815–823 (2016).
Harberts, J. et al. Interfacing human induced pluripotent stem cell-derived neurons with designed nanowire arrays as a future platform for medical applications. Biomater. Sci. 8, 2434–2446 (2020).
Kwon, J. et al. Vertical nanowire electrode array for enhanced neurogenesis of human neural stem cells via intracellular electrical stimulation. Nano Lett. 21, 6343–6351 (2021).
Gautam, V. et al. Engineering highly interconnected neuronal networks on nanowire scaffolds. Nano Lett. 17, 3369–3375 (2017).
Prinz, C. et al. Axonal guidance on patterned free-standing nanowire surfaces. Nanotechnology 19, 345101 (2008).
Bhingardive, V. et al. Nanowire based mechanostimulating platform for tunable activation of natural killer cells. Adv. Funct. Mater. 31, 2103063 (2021).
McWhorter, F. Y. et al. Modulation of macrophage phenotype by cell shape. Proc. Natl Acad. Sci. USA 110, 17253–17258 (2013).
Bhingardive, V. et al. Antibody-functionalized nanowires: a tuner for the activation of T cells. Nano Lett. 21, 4241–4248 (2021).
Wang, J. et al. Physical activation of innate immunity by spiky particles. Nat. Nanotechnol. 13, 1078–1086 (2018).
Le Saux, G. et al. Nanoscale mechanosensing of natural killer cells is revealed by antigen-functionalized nanowires. Adv. Mater. 31, 1805954 (2019).
Arias, S. L. et al. Bacterial envelope damage inflicted by bioinspired nanostructures grown in a hydrogel. ACS Appl. Bio Mater. 3, 7974–7988 (2020).
Bhadra, C. M. et al. Antibacterial titanium nano-patterned arrays inspired by dragonfly wings. Sci. Rep. 5, 16817 (2015).
Hasan, J. et al. Multi-scale surface topography to minimize adherence and viability of nosocomial drug-resistant bacteria. Mater. Des. 140, 332–344 (2018).
Jenkins, J. et al. Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 11, 1626 (2020).
Michalska, M. et al. Tuning antimicrobial properties of biomimetic nanopatterned surfaces. Nanoscale 10, 6639–6650 (2018).
Velic, A. et al. Effects of nanopillar size and spacing on mechanical perturbation and bactericidal killing efficiency. Nanomaterials 11, 2472 (2021).
Zhang, A., Lee, J.-H. & Lieber, C. M. Nanowire-enabled bioelectronics. Nano Today 38, 101135 (2021).
Spira, M. E. & Hai, A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 8, 83–94 (2013).
Mariano, A. et al. Advances in cell-conductive polymer biointerfaces and role of the plasma membrane. Chem. Rev. 122, 4552–4580 (2022).
Hai, A., Shappir, J. & Spira, M. E. Long-term, multisite, parallel, in-cell recording and stimulation by an array of extracellular microelectrodes. J. Neurophysiol. 104, 559–568 (2010).
Hai, A., Shappir, J. & Spira, M. E. In-cell recordings by extracellular microelectrodes. Nat. Methods 7, 200–202 (2010).
Shmoel, N. et al. Multisite electrophysiological recordings by self-assembled loose-patch-like junctions between cultured hippocampal neurons and mushroom-shaped microelectrodes. Sci. Rep. 6, 27110 (2016).
Chen, R., Canales, A. & Anikeeva, P. Neural recording and modulation technologies. Nat. Rev. Mater. 2, 16093 (2017).
McGuire, A. F., Santoro, F. & Cui, B. Interfacing cells with vertical nanoscale devices: applications and characterization. Annu. Rev. Anal. Chem. 11, 101–126 (2018).
Abbott, J. et al. Optimizing nanoelectrode arrays for scalable intracellular electrophysiology. Acc. Chem. Res. 51, 600–608 (2018).
Tian, B. et al. Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science 329, 830–834 (2010).
Fu, T.-M. et al. Sub-10-nm intracellular bioelectronic probes from nanowire–nanotube heterostructures. Proc. Natl Acad. Sci. USA 111, 1259–1264 (2014).
Qing, Q. et al. Free-standing kinked nanowire transistor probes for targeted intracellular recording in three dimensions. Nat. Nanotechnol. 9, 142–147 (2014).
Yoo, J. et al. Long-term intracellular recording of optogenetically-induced electrical activities using vertical nanowire multi electrode array. Sci. Rep. 10, 4279 (2020).
Liu, R. et al. High density individually addressable nanowire arrays record intracellular activity from primary rodent and human stem cell derived neurons. Nano Lett. 17, 2757–2764 (2017).
Staufer, O. et al. Adhesion stabilized en masse intracellular electrical recordings from multicellular assemblies. Nano Lett. 19, 3244–3255 (2019).
Verma, P. & Melosh, N. A. Gigaohm resistance membrane seals with stealth probe electrodes. Appl. Phys. Lett. 97, 033704 (2010).
Keefer, E. W. et al. Carbon nanotube coating improves neuronal recordings. Nat. Nanotechnol. 3, 434–439 (2008).
Hong, G. et al. Mesh electronics: a new paradigm for tissue-like brain probes. Curr. Opin. Neurobiol. 50, 33–41 (2018).
Hong, G. & Lieber, C. M. Novel electrode technologies for neural recordings. Nat. Rev. Neurosci. 20, 330–345 (2019).
Tang, J. et al. Nanowire arrays restore vision in blind mice. Nat. Commun. 9, 786 (2018).
Suyatin, D. B. et al. Nanowire-based electrode for acute in vivo neural recordings in the brain. PLoS ONE 8, e56673 (2013).
Xie, C. et al. Intracellular recording of action potentials by nanopillar electroporation. Nat. Nanotechnol. 7, 185–190 (2012).
Abbott, J. et al. A nanoelectrode array for obtaining intracellular recordings from thousands of connected neurons. Nat. Biomed. Eng. 4, 232–241 (2020).
Abbott, J. et al. CMOS nanoelectrode array for all-electrical intracellular electrophysiological imaging. Nat. Nanotechnol. 12, 460–466 (2017).
Ham, D. et al. Neuromorphic electronics based on copying and pasting the brain. Nat. Electron. 4, 635–644 (2021).
Shokouhi, A.-R. et al. Vertically configured nanostructure-mediated electroporation: a promising route for intracellular regulations and interrogations. Mater. Horiz. 7, 2810–2831 (2020).
Dipalo, M. et al. Intracellular and extracellular recording of spontaneous action potentials in mammalian neurons and cardiac cells with 3D plasmonic nanoelectrodes. Nano Lett. 17, 3932–3939 (2017).
Chen, C. et al. Electrical stimulation as a novel tool for regulating cell behavior in tissue engineering. Biomater. Res. 23, 25 (2019).
Ghezzi, D. et al. A hybrid bioorganic interface for neuronal photoactivation. Nat. Commun. 2, 166 (2011).
Ghezzi, D. et al. A polymer optoelectronic interface restores light sensitivity in blind rat retinas. Nat. Photonics 7, 400–406 (2013).
Mathieson, K. et al. Photovoltaic retinal prosthesis with high pixel density. Nat. Photonics 6, 391–397 (2012).
Parameswaran, R. et al. Optical stimulation of cardiac cells with a polymer-supported silicon nanowire matrix. Proc. Natl Acad. Sci. USA 116, 413–421 (2019).
Rotenberg, M. Y. et al. Silicon nanowires for intracellular optical interrogation with subcellular resolution. Nano Lett. 20, 1226–1232 (2020).
Parameswaran, R. et al. Photoelectrochemical modulation of neuronal activity with free-standing coaxial silicon nanowires. Nat. Nanotechnol. 13, 260–266 (2018).
Han, X. et al. Silicon nanowire-based surface-enhanced Raman spectroscopy endoscope for intracellular pH detection. ACS Appl. Mater. Interfaces 5, 5811–5814 (2013).
Barrios-Rodiles, M. et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005).
Papalexi, E. & Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35–45 (2018).
Wang, Z. et al. High-throughput intracellular biopsy of microRNAs for dissecting the temporal dynamics of cellular heterogeneity. Sci. Adv. 6, eaba4971 (2020).
Xie, K. et al. Profiling microRNAs with associated spatial dynamics in acute tissue slices. ACS Nano 15, 4881–4892 (2021).
Xie, C. et al. Vertical nanopillars for highly localized fluorescence imaging. Proc. Natl Acad. Sci. USA 108, 3894–3899 (2011).
Adolfsson, K. et al. Ingestion of gallium phosphide nanowires has no adverse effect on Drosophila tissue function. Nanotechnology 24, 285101 (2013).
ten Siethoff, L. et al. Molecular motor propelled filaments reveal light-guiding in nanowire arrays for enhanced biosensing. Nano Lett. 14, 737–742 (2014).
Frederiksen, R. S. et al. Modulation of fluorescence signals from biomolecules along nanowires due to interaction of light with oriented nanostructures. Nano Lett. 15, 176–181 (2015).
Verardo, D. et al. Nanowires for biosensing: lightguiding of fluorescence as a function of diameter and wavelength. Nano Lett. 18, 4796–4802 (2018).
Harris, A. K., Wild, P. & Stopak, D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208, 177–179 (1980).
Engler, A. J. et al. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Munevar, S., Wang, Y.-l & Dembo, M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys. J. 80, 1744–1757 (2001).
Kraning-Rush, C. M., Califano, J. P. & Reinhart-King, C. A. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE 7, e32572 (2012).
Koch, T. M. et al. 3D traction forces in cancer cell invasion. PLoS ONE 7, e33476 (2012).
Dembo, M. et al. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys. J. 70, 2008–2022 (1996).
Beningo, K. A., Lo, C.-M. & Wang, Y.-L. Flexible polyacrylamide substrata for the analysis of mechanical interactions at cell-substratum adhesions. Methods Cell Biol. 69, 325–339 (2002).
Pelham, R. J. Jr & Wang, Y.-l High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol. Biol. Cell 10, 935–945 (1999).
Tan, J. L. et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100, 1484–1489 (2003).
Rape, A. D., Guo, W.-H. & Wang, Y.-L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials 32, 2043–2051 (2011).
Trichet, L. et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc. Natl Acad. Sci. USA 109, 6933–6938 (2012).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012).
Miyamoto, S., Akiyama, S. K. & Yamada, K. M. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science 267, 883–885 (1995).
Schäfer, C. et al. One step ahead: role of filopodia in adhesion formation during cell migration of keratinocytes. Exp. Cell Res. 315, 1212–1224 (2009).
Kuo, C. W. et al. Polymeric nanopillar arrays for cell traction force measurements. Electrophoresis 31, 3152–3158 (2010).
Hällström, W. et al. Fifteen-piconewton force detection from neural growth cones using nanowire arrays. Nano Lett. 10, 782–787 (2010).
Li, Z. et al. Quantifying the traction force of a single cell by aligned silicon nanowire array. Nano Lett. 9, 3575–3580 (2009).
Li, Z. et al. Cellular traction forces: a useful parameter in cancer research. Nanoscale 9, 19039–19044 (2017).
Zheng, Q. et al. Dynamic real-time imaging of living cell traction force by piezo-phototronic light nano-antenna array. Sci. Adv. 7, eabe7738 (2021).
Wolfenson, H. et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat. Cell Biol. 18, 33–42 (2016).
da Silva, A. M. et al. Nanowire arrays as force sensors with super-resolved localization position detection: application to optical measurement of bacterial adhesion forces. Small Methods 2, 1700411 (2018).
Kumar, A. R. et al. Materials for improving immune cell transfection. Adv. Mater. 33, 2007421 (2021).
Lestrell, E., O’Brien, C. M., Elnathan, R. & Voelcker, N. H. Vertically aligned nanostructured topographies for human neural stem cell differentiation and neuronal cell interrogation. Adv. Ther. 4, 2100061 (2021).
Schmiderer, L. et al. Efficient and nontoxic biomolecule delivery to primary human hematopoietic stem cells using nanostraws. Proc. Natl Acad. Sci. USA 117, 21267–21273 (2020).
Yosef, N. et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013).
Cao, Y. et al. Universal intracellular biomolecule delivery with precise dosage control. Sci. Adv. 4, eaat8131 (2018).
Dixit, H. G. et al. Massively-parallelized, deterministic mechanoporation for intracellular delivery. Nano Lett. 20, 860–867 (2020).
Elnathan, R. et al. Engineering vertically aligned semiconductor nanowire arrays for applications in the life sciences. Nano Today 9, 172–196 (2014).
Tay, A. The benefits of going small: nanostructures for mammalian cell transfection. ACS Nano 14, 7714–7721 (2020).
Chiappini, C. & Almeida, C. in Semiconducting Silicon Nanowires for Biomedical Applications (ed. Coffer, J. L.) 144–167 (Elsevier, 2014).
Han, S.-W. et al. High-efficiency DNA injection into a single human mesenchymal stem cell using a nanoneedle and atomic force microscopy. Nanomed. Nanotechnol. Biol. Med. 4, 215–225 (2008).
Leprince, L. et al. Dexamethasone electrically controlled release from polypyrrole-coated nanostructured electrodes. J. Mater. Sci. Mater. Med. 21, 925–930 (2010).
Chen, X. et al. High-throughput delivery: a diamond nanoneedle array for potential high-throughput intracellular delivery. Adv. Healthc. Mater. 2, 1065–1065 (2013).
Chiappini, C. et al. Biodegradable porous silicon barcode nanowires with defined geometry. Adv. Funct. Mater. 20, 2231–2239 (2010).
Salonen, J. et al. Mesoporous silicon microparticles for oral drug delivery: loading and release of five model drugs. J. Control. Release 108, 362–374 (2005).
Lee, C. H. et al. Biological lipid membranes for on-demand, wireless drug delivery from thin, bioresorbable electronic implants. NPG Asia Mater. 7, e227–e227 (2015).
Huang, J.-A. et al. On-demand intracellular delivery of single particles in single cells by 3D hollow nanoelectrodes. Nano Lett. 19, 722–731 (2019).
Xu, A. M. et al. Temporally resolved direct delivery of second messengers into cells using nanostraws. Lab Chip 16, 2434–2439 (2016).
Lee, K. et al. Physical delivery of macromolecules using high-aspect ratio nanostructured materials. ACS Appl. Mater. Interfaces 7, 23387–23397 (2015).
Kim, K. & Lee, W. G. Electroporation for nanomedicine: a review. J. Mater. Chem. B 5, 2726–2738 (2017).
Fajrial, A. K. et al. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics 10, 5532 (2020).
Saklayen, N. et al. Intracellular delivery using nanosecond-laser excitation of large-area plasmonic substrates. ACS Nano 11, 3671–3680 (2017).
Wang, Y. et al. High-efficiency cellular reprogramming by nanoscale puncturing. Nano Lett. 20, 5473–5481 (2020).
Chen, Y. et al. Engineering micro–nanomaterials for biomedical translation. Adv. NanoBiomed Res. 1, 2100002 (2021).
Choi, M. et al. Intracellular delivery of bioactive cargos to hard-to-transfect cells using carbon nanosyringe arrays under an applied centrifugal g-force. Adv. Healthc. Mater. 5, 101–107 (2016).
Pan, J. et al. Stimulation of gene transfection by silicon nanowire arrays modified with polyethylenimine. ACS Appl. Mater. Interfaces 6, 14391–14398 (2014).
Chan, M. S. & Lo, P. K. Nanoneedle-assisted delivery of site-selective peptide-functionalized DNA nanocages for targeting mitochondria and nuclei. Small 10, 1255–1260 (2014).
Xu, A. M. et al. Direct intracellular delivery of cell-impermeable probes of protein glycosylation by using nanostraws. ChemBioChem 18, 623–628 (2017).
Shalek, A. K. et al. Nanowire-mediated delivery enables functional interrogation of primary immune cells: application to the analysis of chronic lymphocytic leukemia. Nano Lett. 12, 6498–6504 (2012).
Karra, D. & Dahm, R. Transfection techniques for neuronal cells. J. Neurosci. 30, 6171–6177 (2010).
Kim, H. et al. Bioresorbable, miniaturized porous silicon needles on a flexible water-soluble backing for unobtrusive, sustained delivery of chemotherapy. ACS Nano 14, 7227–7236 (2020).
Tay, A. & Melosh, N. Mechanical stimulation after centrifuge-free nano-electroporative transfection is efficient and maintains long-term T cell functionalities. Small 17, 2103198 (2021).
Fox, C. B. et al. Fabrication of sealed nanostraw microdevices for oral drug delivery. ACS Nano 10, 5873–5881 (2016).
Hebisch, E. et al. Nanostraw-assisted cellular injection of fluorescent nanodiamonds via direct membrane opening. Small 17, 2006421 (2021).
Piret, G., Perez, M.-T. & Prinz, C. N. Neurite outgrowth and synaptophysin expression of postnatal CNS neurons on GaP nanowire arrays in long-term retinal cell culture. Biomaterials 34, 875–887 (2013).
Li, Z. et al. Morphology of living cells cultured on nanowire arrays with varying nanowire densities and diameters. Sci. China Life Sci. 61, 427–435 (2018).
Li, Z. et al. Single cell analysis of proliferation and movement of cancer and normal-like cells on nanowire array substrates. J. Mater. Chem. B 6, 7042–7049 (2018).
Roy, A. R. et al. Exploring cell surface–nanopillar interactions with 3D super-resolution microscopy. ACS Nano 16, 192–210 (2022).
Lin, Z. C. et al. Accurate nanoelectrode recording of human pluripotent stem cell-derived cardiomyocytes for assaying drugs and modeling disease. Microsyst. Nanoeng. 3, 1–7 (2017).
Acknowledgements
R.E. thanks the Australian government (ARC DECRA project number: DE170100021), the Melbourne Centre for Nanofabrication (MCN) in the Victorian Node of the Australian National Fabrication Facility (ANFF), the ANFF-Vic Tech Ambassador Program for Deakin University, Deakin’s School of Medicine and Deakin’s Institute of Frontier Materials. X.X. acknowledges financial support from the National Natural Science Foundation of China (grant no. 32171399) and National Key R&D Program of China (grant no. 2021YFF1200700, 2021YFA0911100). P.S. acknowledges support from the Hong Kong Centre for Cerebro-cardiovascular Health Engineering, funded by the Innovation and Technology Commission of Hong Kong. F.S. acknowledges the support of the European Research Council starting grant BRAIN-ACT no. 949478. C.C. acknowledges the support of the European Research Council starting grant ENBION no. 759577. Y.Z. acknowledges the support of the UK Department for Business, Energy, and Industrial Strategy through the National Measurement System (NMS project, Bioelectronics integrated multifunctional physiological measurement platform) and EPSRC Industrial CASE 2020 (20000128). W.Z. acknowledges the support of the Singapore Ministry of Education (MOE) (W.Z., RG112/20, NGF-2021-10-026 and MOET32020-0001), the Singapore National Research Foundation (W.Z., NRF2019-NRF-ISF003-3292), the Human Frontier Science Program (RGY0088/2021) and the NTU start-up grant. N.H.V. thanks the Australian Research Council for support under the Industrial Transformation Training Centre Scheme (IC170100016 and IC190100026).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Materials thanks Mark Schvartzman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elnathan, R., Barbato, M.G., Guo, X. et al. Biointerface design for vertical nanoprobes. Nat Rev Mater 7, 953–973 (2022). https://doi.org/10.1038/s41578-022-00464-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41578-022-00464-7
This article is cited by
-
Multimodal probing of T-cell recognition with hexapod heterostructures
Nature Methods (2024)
-
Monolithic-to-focal evolving biointerfaces in tissue regeneration and bioelectronics
Nature Chemical Engineering (2024)
-
Electroactive nanoinjection platform for intracellular delivery and gene silencing
Journal of Nanobiotechnology (2023)
-
Regulation of biological processes by intrinsically chiral engineered materials
Nature Reviews Materials (2023)
-
Role of actin cytoskeleton in cargo delivery mediated by vertically aligned silicon nanotubes
Journal of Nanobiotechnology (2022)