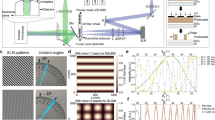
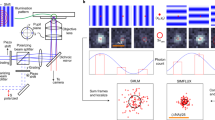
Abstract
Light microscopy is indispensable for analysis of bacterial spatial organization, yet the sizes and shapes of bacterial cells pose unique challenges to imaging. Bacterial cells are not much larger than the diffraction limit of visible light, and many species have cylindrical shapes and so lie flat on microscope coverslips, yielding low-resolution images when observing their short axes. In this protocol, we describe a pair of recently developed methods named VerCINI (vertical cell imaging by nanostructured immobilization) and µVerCINI (microfluidic VerCINI) that greatly increase spatial resolution and image quality for microscopy of the short axes of bacteria. The concept behind both methods is that cells are imaged while confined vertically inside cell traps made from a nanofabricated mold. The mold is a patterned silicon wafer produced in a cleanroom facility using electron-beam lithography and deep reactive ion etching, which takes ~3 h for fabrication and ~12 h for surface passivation. After obtaining a mold, the entire process of making cell traps, imaging cells and processing images can take ~2–12 h, depending on the experiment. VerCINI and µVerCINI are ideal for imaging any process along the short axes of bacterial cells, as they provide high-resolution images without any special requirements for fluorophores or imaging modalities, and can readily be combined with other imaging methods (e.g., STORM). VerCINI can easily be incorporated into existing projects by researchers with expertise in bacteriology and microscopy. Nanofabrication can be either done in-house, requiring specialist facilities, or outsourced based on this protocol.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Source data for all figures presented in the paper are available at figshare (https://doi.org/10.25405/data.ncl.c.5652010.v1).
Code availability
Custom software is available on the Holden lab GitHub page or Zenodo: https://github.com/HoldenLab/VerCINI_nanofab25, https://github.com/HoldenLab/DeepAutoFocus37, https://github.com/HoldenLab/VerciniAnalysisJ38 and https://github.com/HoldenLab/ring-fitting230.
References
Huang, B., Babcock, H. & Zhuang, X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 (2010).
Pasquina-Lemonche, L. et al. The architecture of the Gram-positive bacterial cell wall. Nature 582, 294–297 (2020).
Garner, E. C. et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011).
Domínguez-Escobar, J. et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011).
Bisson-Filho, A. W. et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017).
Yang, X. et al. GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017).
Kirkpatrick, C. L. & Viollier, P. H. Poles apart: prokaryotic polar organelles and their spatial regulation. Cold Spring Harb. Perspect. Biol. 3, a006809–a006809 (2011).
Kuwada, N. J., Traxler, B. & Wiggins, P. A. Genome‐scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle. Mol. Microbiol. 95, 64–79 (2015).
Laloux, G. & Jacobs-Wagner, C. How do bacteria localize proteins to the cell pole? J. Cell Sci. https://doi.org/10.1242/jcs.138628 (2014).
Surovtsev, I. V. & Jacobs-Wagner, C. Subcellular organization: a critical feature of bacterial cell replication. Cell 172, 1271–1293 (2018).
Bowman, G. R., Lyuksyutova, A. I. & Shapiro, L. Bacterial polarity. Curr. Opin. Cell Biol. 23, 71–77 (2011).
Whitley, K. D. et al. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat. Commun. 12, 2448 (2021).
Perez, A. J. et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA 116, 3211–3220 (2019).
Chai, Y., Norman, T., Kolter, R. & Losick, R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 24, 754–765 (2010).
Söderström, B., Chan, H., Shilling, P. J., Skoglund, U. & Daley, D. O. Spatial separation of FtsZ and FtsN during cell division. Mol. Microbiol. 107, 387–401 (2018).
McCausland, J. W. et al. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Commun. 12, 609 (2021).
Trouve, J. et al. Nanoscale dynamics of peptidoglycan assembly during the cell cycle of Streptococcus pneumoniae. Curr. Biol. https://doi.org/10.1016/j.cub.2021.04.041 (2021).
Bakshi, S., Choi, H. & Weisshaar, J. C. The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 6, 636 (2015).
Zhou, Z. et al. The contractile ring coordinates curvature-dependent septum assembly during fission yeast cytokinesis. Mol. Biol. Cell 26, 78–90 (2015).
Thiyagarajan, S., Munteanu, E. L., Arasada, R., Pollard, T. D. & O’Shaughnessy, B. The fission yeast cytokinetic contractile ring regulates septum shape and closure. J. Cell Sci. https://doi.org/10.1242/jcs.166926 (2015).
Wollrab, V., Thiagarajan, R., Wald, A., Kruse, K. & Riveline, D. Still and rotating myosin clusters determine cytokinetic ring constriction. Nat. Commun. 7, 11860 (2016).
Strahl, H. et al. Transmembrane protein sorting driven by membrane curvature. Nat. Commun. 6, (2015).
Briegel, A. et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry 53, 1575–1585 (2014).
Deshpande, S. & Dekker, C. On-chip microfluidic production of cell-sized liposomes. Nat. Protoc. 13, 856–874 (2018).
Whitley, K. VerCINI_nanofab. Zenodo https://doi.org/10.5281/ZENODO.5548030 (2021).
de Jong, I. G., Beilharz, K., Kuipers, O. P. & Veening, J.-W. Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J. Vis. Exp. 53, e3145 (2011).
Ellefsen, K. L., Dynes, J. L. & Parker, I. Spinning-spot shadowless TIRF microscopy. PLoS ONE 10, e0136055 (2015).
McGorty, R., Kamiyama, D. & Huang, B. Active microscope stabilization in three dimensions using image correlation. Opt. Nanoscopy 2, 3 (2013).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Holden, S. & Whitley, K. HoldenLab/ring-fitting2: v1.1.2.1. Zenodo https://doi.org/10.5281/ZENODO.4570259 (2021).
Luisier, F., Vonesch, C., Blu, T. & Unser, M. Fast interscale wavelet denoising of Poisson-corrupted images. Signal Process 90, 415–427 (2010).
Thevenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process 7, 27–41 (1998).
Jakobs, M. A., Dimitracopoulos, A. & Franze, K. KymoButler, a deep learning software for automated kymograph analysis. eLife 8, e42288 (2019).
Wang, P. et al. Robust growth of Escherichia coli. Curr. Biol. 20, 1099–1103 (2010).
Tam, J. et al. A microfluidic platform for correlative live-cell and super-resolution microscopy. PloS ONE 9, e115512 (2014).
Almada, P. et al. Automating multimodal microscopy with NanoJ-Fluidics. Nat. Commun. 10, 1223 (2019).
Whitley, K. D. et al. DeepAutoFocus. Zenodo https://doi.org/10.5281/ZENODO.4573121 (2021).
Holden, S. HoldenLab/VerciniAnalysisJ: v1.1.1. Zenodo https://doi.org/10.5281/ZENODO.4617277 (2021).
Erratum for the Report: "Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division" by A. W. Bisson-Filho, Y.-P. Hsu, G. R. Squyres, E. Kuru, F. Wu, C. Jukes, Y. Sun, C. Dekker, S. Holden, M. S. VanNieuwenhze, Y. V. Brun, E. C. GarnerErratum for the Report: “Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division”. Science 367, eaba6311 (2020).
Tinevez, J.-Y. et al. TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Acknowledgements
We acknowledge S. Pud (TU Delft; now at Twente) for help with wafer design and nanofabrication, S. Deshpande (TU Delft; now at Wageningen) for help with nanofabrication and soft lithography, M. Zuiddam (TU Delft) for help with wafer etching, H. Strahl (Newcastle) for strains and helpful discussions, and J. Fritzsche (ConScience AB, Sweden) for helpful discussions. S.H., K.D.W., C.J. and S.M. acknowledge funding support by a Wellcome Trust & Royal Society Sir Henry Dale Fellowship [206670/Z/17/Z]. C.D. acknowledges funding support by from the ERC Advanced Grants [883684] and [669598], and the NanoFront and BaSyC programs. S.M. is supported by a UK Biotechnology and Biological Sciences Research Council doctoral studentship.
Author information
Authors and Affiliations
Contributions
K.D.W., S.M. and C.J. performed the experiments. K.D.W., S.M., S.H. and C.D. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Cecile Morlot and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Bisson-Filho, A. W. et al. Science 355, 739–743 (2017): https://doi.org/10.1126/science.aak9973
Perez, A. J. et al. Proc. Natl. Acad. Sci. USA 116, 3211–3220 (2019): https://doi.org/10.1073/pnas.1816018116
Whitley, K. D. et al. Nat. Commun. 12, 2448 (2021): https://doi.org/10.1038/s41467-021-22526-0
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Methods.
Supplementary Video 1
Suboptimal cell trapping visualized by bright-field microscopy. B. subtilis PY79 cells improperly trapped in microhole arrays where microhole width (>1.1 µm) is too large to immobilize most cells. Lateral diffusive motion in the holes (wobbling) can be observed in ~50% of trapped cells. The video shows acceptable but not excellent cell loading efficiency. A few cells can be seen sitting on top of rather than within microholes, likely those that did not wash off during sample preparation. Video was recorded at 100 Hz and plays in real time. Scale bar, 10 µm.
Rights and permissions
About this article
Cite this article
Whitley, K.D., Middlemiss, S., Jukes, C. et al. High-resolution imaging of bacterial spatial organization with vertical cell imaging by nanostructured immobilization (VerCINI). Nat Protoc 17, 847–869 (2022). https://doi.org/10.1038/s41596-021-00668-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00668-1
This article is cited by
-
Peptidoglycan synthesis drives a single population of septal cell wall synthases during division in Bacillus subtilis
Nature Microbiology (2024)
-
DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.