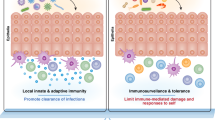
Abstract
Regulatory T cells (Treg cells) are key players in ensuring a peaceful coexistence with microorganisms and food antigens at intestinal borders. Startling new information has appeared in recent years on their diversity, the importance of the transcription factor FOXP3, how T cell receptors influence their fate and the unexpected and varied cellular partners that influence Treg cell homeostatic setpoints. We also revisit some tenets, maintained by the echo chambers of Reviews, that rest on uncertain foundations or are a subject of debate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tanoue, T., Atarashi, K. & Honda, K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16, 295–309 (2016).
Russler-Germain, E. V., Rengarajan, S. & Hsieh, C. S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 10, 1375–1386 (2017).
Jacobse, J. et al. Intestinal regulatory T cells as specialized tissue-restricted immune cells in intestinal immune homeostasis and disease. Front. Immunol. 12, 716499 (2021).
Traxinger, B. R., Richert-Spuhler, L. E. & Lund, J. M. Mucosal tissue regulatory T cells are integral in balancing immunity and tolerance at portals of antigen entry. Mucosal Immunol. 15, 398–407 (2022).
van der Veeken, J. et al. The transcription factor Foxp3 shapes regulatory T cell identity by tuning the activity of trans-acting intermediaries. Immunity 53, 971–984 (2020).
Panduro, M., Benoist, C. & Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 34, 609–633 (2016).
Cosovanu, C. & Neumann, C. The many functions of Foxp3+ regulatory T cells in the intestine. Front. Immunol. 11, 600973 (2020).
Roncarolo, M. G. et al. The biology of T regulatory type 1 cells and their therapeutic application in immune-mediated diseases. Immunity 49, 1004–1019 (2018).
Joeris, T. et al. Intestinal cDC1 drive cross-tolerance to epithelial-derived antigen via induction of FoxP3+CD8+ Tregs. Sci. Immunol. 6, eabd3774 (2021).
Dispirito, J. R. et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 3, eaat5861 (2018).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011).
Faith, J. J. et al. Identifying gut microbe–host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra11 (2014).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Kim, K. S. et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016).
Hong, S.-W. et al. Immune tolerance of food is mediated by layers of CD4+ T cell dysfunction. Nature 607, 762–768 (2022). This study shows that exposure to food antigens causes cognate CD4+ T cells to acquire diverse hyporesponsive TH cell and Treg cell phenotypes.
Gélineau, A. Dietary fibers benefits on glucose homeostasis require type 2 conventional dendritic cells in mice fed a high-fat diet. Preprint at bioRxiv https://doi.org/10.1101/2023.04.19.537402 (2023).
Duhen, T. et al. Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector TH cells. Blood 119, 4430–4440 (2012).
Wohlfert, E. A. et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121, 4503–4515 (2011).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Neumann, C. et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host–microbiota homeostasis. Nat. Immunol. 20, 471–481 (2019). This paper suggests that intestinal Treg cells may control the composition of the microbiota.
Yang, B. H. et al. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal. Immunol. 9, 444–457 (2016).
Ohnmacht, C. et al. The microbiota regulates type 2 immunity through RORγ+ T cells. Science 349, 989–993 (2015).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015). Together with Ohnmacht et al. (2015), this study provides the first description of microbial influence on setting the RORγ+ Treg cell phenotype.
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Wheaton, J. D., Yeh, C. H. & Ciofani, M. Cutting Edge: c-Maf is required for regulatory T cells to adopt RORγt+ and follicular phenotypes. J. Immunol. 199, 3931–3936 (2017).
Krzyzanowska, A. K. et al. Zbtb20 identifies and controls a thymus-derived population of regulatory T cells that play a role in intestinal homeostasis. Sci. Immunol. 7, eabf3717 (2022).
Levine, A. G. et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017).
Britton, G. J. et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut TH17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity 50, 212–224 (2019).
van der Veeken, J. et al. Genetic tracing reveals transcription factor Foxp3-dependent and Foxp3-independent functionality of peripherally induced Treg cells. Immunity 55, 1173–1184 (2022). Using an astute fate mapping strategy, this study shows that most RORγ+ Treg cells are pTreg cells (at least in the context of reversion from RORγ+ Treg cell deprivation).
Chowdhary, K., et al. An interwoven network of transcription factors, with divergent influences from FoxP3, underlies Treg diversity. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2023.05.18.541358v1 (2023). This preprint confirms a key observation from van der Veeken et al. (2022) that RORγ+ Treg cells in the intestine function independently of FOXP3.
Sujino, T. et al. Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science 352, 1581–1586 (2016).
Betts, C. B. et al. Mucosal immunity in the female murine mammary gland. J. Immunol. 201, 734–746 (2018).
Geva-Zatorsky, N. et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943 (2017).
Hanna, B. S. et al. The gut microbiota promotes distal tissue regeneration via RORγ+ regulatory T cell emissaries. Immunity 56, 829–846 (2023). This work shows that intestinal RORγ+ Treg cells can partake in regulatory phenomena in other organs (injured muscle and liver) during inflammation.
Zemmour, D. et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19, 291–301 (2018).
Pratama, A., Schnell, A., Mathis, D. & Benoist, C. Developmental and cellular age direct conversion of CD4+ T cells into RORγ+ or Helios+ colon Treg cells. J. Exp. Med. 217, e20190428 (2020).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504 (2019).
Delacher, M. et al. Single-cell chromatin accessibility landscape identifies tissue repair program in human regulatory T cells. Immunity 54, 702–720 (2021).
Sassone-Corsi, M. et al. Sequestration of gut pathobionts in intraluminal casts, a mechanism to avoid dysregulated T cell activation by pathobionts. Proc. Natl Acad. Sci. USA 119, e2209624119 (2022).
London, M. et al. Stepwise chromatin and transcriptional acquisition of an intraepithelial lymphocyte program. Nat. Immunol. 22, 449–459 (2021).
Thornton, A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010).
Weiss, J. M. et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209, 1723–1742 (2012).
Knoop, K. A. et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci. Immunol. 2, eaao1314 (2017). Among the extreme diversity of signals involved in Treg cell homeostasis, this work reveals that establishment of the Treg cell setpoint depends on antigen exposure during the critical window of postnatal development.
Abdel-Gadir, A. et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 25, 1164–1174 (2019).
Al Nabhani, Z. et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288 (2019).
Ramanan, D. et al. Homeostatic, repertoire and transcriptional relationships between colon T regulatory cell subsets. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2023.05.17.541199v (2023).
Hemmers, S., Schizas, M. & Rudensky, A. Y. Treg cell-intrinsic requirements for ST2 signaling in health and neuroinflammation. J. Exp. Med. 218, e20201234 (2021).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Morton, A. M. et al. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc. Natl Acad. Sci. USA 111, 6696–6701 (2014).
Nakanishi, Y. et al. Regulatory T cells with superior immunosuppressive capacity emigrate from the inflamed colon to draining lymph nodes. Mucosal. Immunol. 11, 437–448 (2018).
Galván-Peña, S. et al. A dynamic atlas of immunocyte migration from the gut. Preprint at bioRxiv https://doi.org/10.1101/2022.11.16.516757 (2022).
Knoop, K. A. et al. Synchronization of mothers and offspring promotes tolerance and limits allergy. Jci. Insight 5, e137943 (2020).
Ramanan, D. et al. An immunologic mode of multigenerational transmission governs a gut Treg setpoint. Cell 181, 1276–1290 (2020). This work demonstrates that setpoints of intestinal Treg cell populations are transmitted traits, genetically and with a surprising maternal multi-generational component.
Romagnoli, P., Tellier, J. & van Meerwijk, J. P. Genetic control of thymic development of CD4+CD25+FoxP3+ regulatory T lymphocytes. Eur. J. Immunol. 35, 3525–3532 (2005).
Depis, F., Kwon, H. K., Mathis, D. & Benoist, C. Unstable FoxP3+ T regulatory cells in NZW mice. Proc. Natl Acad. Sci. USA 113, 1345–1350 (2016).
Cong, Y. et al. A dominant, coordinated T regulatory cell–IgA response to the intestinal microbiota. Proc. Natl Acad. Sci. USA 106, 19256–19261 (2009).
Kawamoto, S. et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320 (2018).
Campbell, C. et al. Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257 (2018).
Bhattacharjee, A. et al. Environmental enteric dysfunction induces regulatory T cells that inhibit local CD4+ T cell responses and impair oral vaccine efficacy. Immunity 54, 1745–1757 (2021).
Blatner, N. R. et al. Expression of RORγt marks a pathogenic regulatory T cell subset in human colon cancer. Sci. Transl. Med. 4, 164ra159 (2012).
Osman, A. et al. TCF-1 controls Treg cell functions that regulate inflammation, CD8+ T cell cytotoxicity and severity of colon cancer. Nat. Immunol. 22, 1152–1162 (2021).
Wang, Y., Su, M. A. & Wan, Y. Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348 (2011).
Yang, J. et al. Rorc restrains the potency of ST2+ regulatory T cells in ameliorating intestinal graft-versus-host disease. Jci. Insight 4, e122014 (2019).
Fulton, L. M. et al. Attenuation of acute graft-versus-host disease in the absence of the transcription factor RORγt. J. Immunol. 189, 1765–1772 (2012).
Maywald, R. L. et al. IL-33 activates tumor stroma to promote intestinal polyposis. Proc. Natl Acad. Sci. USA 112, E2487–E2496 (2015).
Syed Khaja, A. S. et al. Intratumoral FoxP3+Helios+ regulatory T cells upregulating immunosuppressive molecules are expanded in human colorectal cancer. Front. Immunol. 8, 619 (2017).
Pastille, E. et al. The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer. Mucosal Immunol. 12, 990–1003 (2019).
Povoleri, G. A. M. et al. Human retinoic acid-regulated CD161+ regulatory T cells support wound repair in intestinal mucosa. Nat. Immunol. 19, 1403–1414 (2018).
James, K. R. et al. Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353 (2020).
Quandt, J. et al. Wnt-β-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression. Nat. Immunol. 22, 471–484 (2021).
Kullberg, M. C. et al. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J. Exp. Med. 196, 505–515 (2002).
Ostman, S. et al. Impaired regulatory T cell function in germ-free mice. Eur. J. Immunol. 36, 2336–2346 (2006).
Ishikawa, H. et al. Effect of intestinal microbiota on the induction of regulatory CD25+CD4+ T cells. Clin. Exp. Immunol. 153, 127–135 (2008).
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Liu, Y. et al. Lactobacillus reuteri DSM 17938 changes the frequency of Foxp3+ regulatory T cells in the intestine and mesenteric lymph node in experimental necrotizing enterocolitis. PLoS ONE 8, e56547 (2013).
Tang, C. et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe 18, 183–197 (2015).
Kuczma, M. P. et al. Commensal epitopes drive differentiation of colonic Tregs. Sci. Adv. 6, eaaz3186 (2020).
Round, J. L. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Dasgupta, S. et al. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 15, 413–423 (2014).
Verma, R. et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3+ regulatory T cells. Sci. Immunol. 3, eaat6975 (2018).
Finney, C. A., Taylor, M. D., Wilson, M. S. & Maizels, R. M. Expansion and activation of CD4+CD25+ regulatory T cells in Heligmosomoides polygyrus infection. Eur. J. Immunol. 37, 1874–1886 (2007).
Johnston, C. J. C. et al. A structurally distinct TGF-β mimic from an intestinal helminth parasite potently induces regulatory T cells. Nat. Commun. 8, 1741 (2017).
Tao, R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307 (2007).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Kim, S. V. et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340, 1456–1459 (2013).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019). Together with Campbell et al. (2020) and Song et al. (2020), this work moves towards the importance of bile acid metabolites in modulating intestinal T cells (regulatory or conventional), and also highlights the complexity of these effects.
Föhse, L. et al. High TCR diversity ensures optimal function and homeostasis of Foxp3+ regulatory T cells. Eur. J. Immunol. 41, 3101–3113 (2011).
Cebula, A. et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 497, 258–262 (2013).
Solomon, B. D. & Hsieh, C. S. Antigen-specific development of mucosal Foxp3+RORγ+ T cells from regulatory T cell precursors. J. Immunol. 197, 3512–3519 (2016).
Chai, J. N. et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2, eaal5068 (2017).
Wegorzewska, M. M. et al. Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol. 4, eaau9079 (2019).
Bousbaine, D. et al. A conserved Bacteroidetes antigen induces anti-inflammatory intestinal T lymphocytes. Science 377, 660–666 (2022).
Ansaldo, E. & Belkaid, Y. How microbiota improve immunotherapy. Science 373, 966–967 (2021).
Kuczma, M. P. et al. Self and microbiota-derived epitopes induce CD4+ T cell anergy and conversion into CD4+Foxp3+ regulatory cells. Mucosal. Immunol. 14, 443–454 (2021).
Nutsch, K. et al. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep. 17, 206–220 (2016).
Russler-Germain, E. V. et al. Commensal Cryptosporidium colonization elicits a cDC1-dependent TH1 response that promotes intestinal homeostasis and limits other infections. Immunity 54, 2547–2564 (2021).
Liu, Z. et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015).
Kretschmer, K. et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6, 1219–1227 (2005).
Apostolou, I. et al. Peripherally induced Treg: mode, stability, and role in specific tolerance. J. Clin. Immunol. 28, 619–624 (2008).
Sun, C. M. et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 Treg cells via retinoic acid. J. Exp. Med. 204, 1775–1785 (2007).
Feuerer, M. et al. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. Immunity 31, 654–664 (2009).
Thorstenson, K. M. & Khoruts, A. Generation of anergic and potentially immunoregulatory CD25+CD4 T cells in vivo after induction of peripheral tolerance with intravenous or oral antigen. J. Immunol. 167, 188–195 (2001).
Coombes, J. L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Yadav, M. et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 209, 1713–1719 (2012).
Szurek, E. et al. Differences in expression level of Helios and Neuropilin-1 do not distinguish thymus-derived from extrathymically-induced CD4+Foxp3+ regulatory T cells. PLoS ONE 10, e0141161 (2015).
Akimova, T. et al. Helios expression is a marker of T cell activation and proliferation. PLoS. ONE 6, e24226 (2011).
Gottschalk, R. A., Corse, E. & Allison, J. P. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J. Immunol. 188, 976–980 (2012).
Haribhai, D. et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity 35, 109–122 (2011).
Kim, B.-S. et al. Generation of RORγt+ antigen-specific T regulatory 17 cells from Foxp3+ precursors in autoimmunity. Cell Rep. 21, 195–207 (2017).
Yang, J. et al. Thymus-derived Foxp3+ regulatory T cells upregulate RORγt expression under inflammatory conditions. J. Mol. Med. 96, 1387–1394 (2018). By showing that tTreg cells can express RORγ upon activation, this study questions the relevance of equating transcription factor profile with Treg cell origin.
Josefowicz, S. Z. et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012).
Holohan, D. R., Van, G. F. & Bluestone, J. A. Thymically-derived Foxp3+ regulatory T cells are the primary regulators of type 1 diabetes in the non-obese diabetic mouse model. PLoS ONE 14, e0217728 (2019).
Muñoz-Rojas, A. R. & Mathis, D. Tissue regulatory T cells: regulatory chameleons. Nat. Rev. Immunol. 21, 597–611 (2021).
Dikiy, S. et al. Terminal differentiation and persistence of effector regulatory T cells essential for the prevention of intestinal inflammation. Preprint at bioRxiv https://www.biorxiv.org/node/2585666 (2022).
Li, C. et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-Treg phenotype. Cell 174, 285–299 (2018).
Delacher, M. et al. Precursors for nonlymphoid-tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity 52, 295–312 (2020).
Li, C. et al. PPARγ marks splenic precursors of multiple nonlymphoid-tissue Treg compartments. Proc. Natl Acad. Sci. USA 118, e2025197118 (2021).
Yissachar, N. et al. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168, 1135–1148 (2017).
Yan, Y. et al. Interleukin-6 produced by enteric neurons regulates the number and phenotype of microbe-responsive regulatory T cells in the gut. Immunity 54, 499–513 (2021).
Sumida, T. et al. Activated β-catenin in Foxp3+ regulatory T cells links inflammatory environments to autoimmunity. Nat. Immunol. 19, 1391–1402 (2018). This paper identifies an important signalling pathway for Treg cells in inflammatory contexts.
Keerthivasan, S. et al. β-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci. Transl. Med. 6, 225ra28 (2014).
Yoshida, H. et al. The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912 (2019).
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008).
Rudra, D. et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13, 1010–1019 (2012).
Kwon, H. K., Chen, H. M., Mathis, D. & Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 18, 1238–1248 (2017).
Apetoh, L. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11, 854–861 (2010).
Gandhi, R. et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3+ regulatory T cells. Nat. Immunol. 11, 846–853 (2010).
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009).
Wheaton, J. D. & Ciofani, M. JunB controls intestinal effector programs in regulatory T cells. Front. Immunol. 11, 444 (2020).
Hayatsu, N. et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity 47, 268–283 (2017).
Ogawa, C. et al. Blimp-1 functions as a molecular switch to prevent inflammatory activity in Foxp3+RORγ+ regulatory T cells. Cell Rep. 25, 19–28 (2018).
Cretney, E. et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12, 304–311 (2011).
Garg, G. et al. Blimp1 prevents methylation of Foxp3 and loss of regulatory T cell identity at sites of inflammation. Cell Rep. 26, 1854–1868 (2019).
Basu, J. et al. Essential role of a ThPOK autoregulatory loop in the maintenance of mature CD4+ T cell identity and function. Nat. Immunol. 22, 969–982 (2021).
Gavin, M. A. et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775 (2007).
Lin, W. et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8, 359–368 (2007).
Charbonnier, L. M. et al. Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat. Immunol. 20, 1208–1219 (2019). This work is an important update on Treg cell ‘wannabes’, Treg-like cells devoid of FOXP3 expression.
Otsubo, K. et al. Identification of FOXP3-negative regulatory T-like (CD4+CD25+CD127low) cells in patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Clin. Immunol. 141, 111–120 (2011).
Boldt, A. et al. Differences in FOXP3 and CD127 expression in Treg-like cells in patients with IPEX syndrome. Clin. Immunol. 153, 109–111 (2014).
Zemmour, D. et al. Single-cell analysis of FOXP3 deficiencies in humans and mice unmasks intrinsic and extrinsic CD4+ T cell perturbations. Nat. Immunol. 22, 607–619 (2021).
Wan, Y. Y. & Flavell, R. A. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007).
Williams, L. M. & Rudensky, A. Y. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 (2007).
Mucida, D. et al. Reciprocal TH-17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007).
Welty, N. E. et al. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J. Exp. Med. 210, 2011–2024 (2013).
Worthington, J. J., Czajkowska, B. I., Melton, A. C. & Travis, M. A. Intestinal dendritic cells specialize to activate transforming growth factor-β and induce Foxp3+ regulatory T cells via integrin αvβ8. Gastroenterology 141, 1802–1812 (2011).
Veenbergen, S. et al. Colonic tolerance develops in the iliac lymph nodes and can be established independent of CD103+ dendritic cells. Mucosal. Immunol. 9, 894–906 (2016).
Esterhazy, D. et al. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral Treg cells and tolerance. Nat. Immunol. 17, 545–555 (2016).
Russler-Germain, E. V. et al. Gut Helicobacter presentation by multiple dendritic cell subsets enables context-specific regulatory T cell generation. eLife 10, e54792 (2021).
Kedmi, R. et al. A RORγ+ cell instructs gut microbiota-specific Treg cell differentiation. Nature 610, 737–743 (2022).
Akagbosu, B. et al. Novel antigen presenting cell imparts Treg-dependent tolerance to gut microbiota. Nature 610, 752–760 (2022).
Lyu, M. et al. ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature 610, 744–751 (2022). Together with Kedmi et al. (2022) and Akagbosu et al. (2022), this study reports the existence of a new subset of RORγ+ intestinal APCs involved in pTreg cell differentiation, but with differing views on the actual identity of the cell or cells.
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Mazzini, E., Massimiliano, L., Penna, G. & Rescigno, M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity 40, 248–261 (2014).
Kim, M., Hill, A. A., Wu, W. J. & Diehl, G. E. Intestinal microbes direct CX3CR1+ cells to balance intestinal immunity. Gut Microbes 10, 540–546 (2019).
Bauche, D. et al. LAG3+ regulatory T cells restrain interleukin-23-producing CX3CR1+ gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity 49, 342–352 (2018).
Fallegger, A. et al. TGF-β production by eosinophils drives the expansion of peripherally induced neuropilin–RORγ+ regulatory T-cells during bacterial and allergen challenge. Mucosal. Immunol. 15, 504–514 (2022).
Mortha, A. et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343, 1249288 (2014).
Zhou, L. et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019).
Deng, T. et al. ILC3-derived OX40L is essential for homeostasis of intestinal Tregs in immunodeficient mice. Cell Mol. Immunol. 17, 163–177 (2020).
Wang, J. et al. Single-cell multiomics defines tolerogenic extrathymic Aire-expressing populations with unique homology to thymic epithelium. Sci. Immunol. 6, eabl5053 (2021).
Ngo Thi Phuong, N. et al. IL-33 drives expansion of type 2 innate lymphoid cells and regulatory T cells and protects mice from severe, acute colitis. Front. Immunol. 12, 669787 (2021).
He, Z. et al. Epithelial-derived IL-33 promotes intestinal tumorigenesis in ApcMin/+ mice. Sci. Rep. 7, 5520 (2017).
Siede, J. et al. IL-33 receptor-expressing regulatory T cells are highly activated, TH2 biased and suppress CD4 T cell proliferation through IL-10 and TGFβ release. PLoS ONE 11, e0161507 (2016).
Harrison, O. J. et al. Epithelial-derived IL-18 regulates TH17 cell differentiation and Foxp3+ Treg cell function in the intestine. Mucosal. Immunol. 8, 1226–1236 (2015).
Nakahashi-Oda, C. et al. Apoptotic epithelial cells control the abundance of Treg cells at barrier surfaces. Nat. Immunol. 17, 441–450 (2016).
Veiga-Fernandes, H. & Mucida, D. Neuro-immune interactions at barrier surfaces. Cell 165, 801–811 (2016).
Jacobson, A., Yang, D., Vella, M. & Chiu, I. M. The intestinal neuro-immune axis: crosstalk between neurons, immune cells, and microbes. Mucosal. Immunol. 14, 555–565 (2021).
Cardoso, V. et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017).
Moriyama, S. et al. β2-Adrenergic receptor-mediated negative regulation of group 2 innate lymphoid cell responses. Science 359, 1056–1061 (2018).
Xu, H. et al. Transcriptional atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity 51, 696–708 (2019).
Chu, C. et al. The ChAT–acetylcholine pathway promotes group 2 innate lymphoid cell responses and anti-helminth immunity. Sci. Immunol. 6, eabe3218 (2021).
Talbot, J. et al. Feeding-dependent VIP neuron–ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580 (2020).
Seillet, C. et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 21, 168–177 (2020).
Muller, P. A. et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158, 300–313 (2014).
Gabanyi, I. et al. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell 164, 378–391 (2016).
Teratani, T. et al. The liver–brain–gut neural arc maintains the Treg cell niche in the gut. Nature 585, 591–596 (2020).
Matheis, F. et al. Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell 180, 64–78 (2020).
Abbas, A. K. et al. Regulatory T cells: recommendations to simplify the nomenclature. Nat. Immunol. 14, 307–308 (2013).
Chen, W. et al. Conversion of peripheral CD4+CD25– naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Hill, J. A. et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi cells. Immunity 29, 758–770 (2008).
Nolting, J. et al. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J. Exp. Med. 206, 2131–2139 (2009).
Mucida, D. et al. Retinoic acid can directly promote TGF-β-mediated Foxp3+ Treg cell conversion of naive T cells. Immunity 30, 471–472 (2009).
Rahim, M. M. et al. Ly49 receptors: innate and adaptive immune paradigms. Front. Immunol. 5, 145 (2014).
Thangavelu, G. et al. Retinoic acid signaling acts as a rheostat to balance Treg function. Cell Mol. Immunol. 19, 820–833 (2022).
Zhou, X. et al. Cutting edge: all-trans retinoic acid sustains the stability and function of natural regulatory T cells in an inflammatory milieu. J. Immunol. 185, 2675–2679 (2010).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Schlenner, S. M. et al. Smad3 binding to the Foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J. Exp. Med. 209, 1529–1535 (2012).
Xu, H. et al. Arkadia-SKI/SnoN signaling differentially regulates TGF-β-induced iTreg and TH17 cell differentiation. J. Exp. Med. 218, e20210777 (2021).
Oh, S. A. et al. Foxp3-independent mechanism by which TGF-β controls peripheral T cell tolerance. Proc. Natl Acad. Sci. USA 114, E7536–E7544 (2017).
Li, W. et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377 (2021).
Koch, M. A. et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016).
Lim, A. I. et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science 373, eabf3002 (2021).
Kim, E. et al. Maternal gut bacteria drive intestinal inflammation in offspring with neurodevelopmental disorders by altering the chromatin landscape of CD4+ T cells. Immunity 55, 145–158 (2022).
Acknowledgements
The authors thank B. Blazar, C. Brown, J. Faith, M. Fischbach, C. S. Hsieh, S. Josefowicz, K. Kretschmer, M. O. Li, D. Littman, A. Rudensky and G. Thangavelu for insightful discussions and/or suggestions or corrections on the draft. Relevant work in the laboratory was supported by US National Institutes of Health (NIH) grants AI125603 and AI150686.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks D. Mucida, H. Ohno and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ramanan, D., Pratama, A., Zhu, Y. et al. Regulatory T cells in the face of the intestinal microbiota. Nat Rev Immunol 23, 749–762 (2023). https://doi.org/10.1038/s41577-023-00890-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-023-00890-w
This article is cited by
-
Impaired immune tolerance mediated by reduced Tfr cells in rheumatoid arthritis linked to gut microbiota dysbiosis and altered metabolites
Arthritis Research & Therapy (2024)
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)
-
Dubosiella newyorkensis modulates immune tolerance in colitis via the L-lysine-activated AhR-IDO1-Kyn pathway
Nature Communications (2024)
-
γδ T cells: origin and fate, subsets, diseases and immunotherapy
Signal Transduction and Targeted Therapy (2023)