Abstract
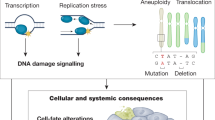
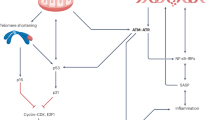
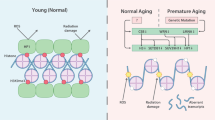
Genomic instability is an important driver of ageing. The accumulation of DNA damage is believed to contribute to ageing by inducing cell death, senescence and tissue dysfunction. However, emerging evidence shows that inflammation is another major consequence of DNA damage. Inflammation is a hallmark of ageing and the driver of multiple age-related diseases. Here, we review the evidence linking DNA damage, inflammation and ageing, highlighting how premature ageing syndromes are associated with inflammation. We discuss the mechanisms by which DNA damage induces inflammation, such as through activation of the cGAS–STING axis and NF-κB activation by ATM. The triggers for activation of these signalling cascades are the age-related accumulation of DNA damage, activation of transposons, cellular senescence and the accumulation of persistent R-loops. We also discuss how epigenetic changes triggered by DNA damage can lead to inflammation and ageing via redistribution of heterochromatin factors. Finally, we discuss potential interventions against age-related inflammation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013). This article presents a comprehensive summary of the molecular characteristics of the ageing process.
Brzostek-Racine, S., Gordon, C., Van Scoy, S. & Reich, N. C. The DNA damage response induces IFN. J. Immunol. 187, 5336–5345 (2011).
Kondo, T. et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl Acad. Sci. Usa. 110, 2969–2974 (2013).
Hartlova, A. et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015).
Sistigu, A. et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014).
Li, T. & Chen, Z. J. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).
van Vugt, M. & Parkes, E. E. When breaks get hot: inflammatory signaling in BRCA1/2-mutant cancers. Trends Cancer 8, 174–189 (2022).
Reislander, T., Groelly, F. J. & Tarsounas, M. DNA damage and cancer immunotherapy: a STING in the tale. Mol. Cell 80, 21–28 (2020).
Fenech, M. et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26, 125–132 (2011).
Erdal, E., Haider, S., Rehwinkel, J., Harris, A. L. & McHugh, P. J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 31, 353–369 (2017).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Krupina, K., Goginashvili, A. & Cleveland, D. W. Causes and consequences of micronuclei. Curr. Opin. Cell Biol. 70, 91–99 (2021).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). This paper for the first time identifies cGAS as a cytosolic DNA sensor.
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012).
Abe, T. & Barber, G. N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 (2014).
Iwanaszko, M. & Kimmel, M. NF-κB and IRF pathways: cross-regulation on target genes promoter level. BMC Genomics 16, 307 (2015).
Ho, S. S. et al. The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity 44, 1177–1189 (2016).
Herzner, A. M. et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16, 1025–1033 (2015).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015). This paper describes the activation of NF-κB by DNA damage independently of cGAS.
Hinz, M. et al. A cytoplasmic ATM–TRAF6–cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol. Cell 40, 63–74 (2010).
Fang, L. et al. ATM regulates NF-κB-dependent immediate-early genes via RelA Ser 276 phosphorylation coupled to CDK9 promoter recruitment. Nucleic Acids Res. 42, 8416–8432 (2014).
d’Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008).
Dunphy, G. et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5 (2018).
Niedernhofer, L. J., Bohr, V. A., Sander, M. & Kraemer, K. H. Xeroderma pigmentosum and other diseases of human premature aging and DNA repair: molecules to patients. Mech. Ageing Dev. 132, 340–347 (2011).
Moraes, M. C., Neto, J. B. & Menck, C. F. DNA repair mechanisms protect our genome from carcinogenesis. Front. Biosci. 17, 1362–1388 (2012).
Cleaver, J. E. Defective repair replication of DNA in xeroderma pigmentosum. Nature 218, 652–656 (1968).
Cleaver, J. E. Xeroderma pigmentosum: a human disease in which an initial stage of DNA repair is defective. Proc. Natl Acad. Sci. Usa. 63, 428–435 (1969).
Sijbers, A. M. et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86, 811–822 (1996).
Niedernhofer, L. J. et al. The structure-specific endonuclease Ercc1–Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24, 5776–5787 (2004).
Klein Douwel, D. et al. XPF–ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 54, 460–471 (2014).
Niedernhofer, L. J. et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043 (2006).
Karakasilioti, I. et al. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 18, 403–415 (2013).
Kim, D. E. et al. Deficiency in the DNA repair protein ERCC1 triggers a link between senescence and apoptosis in human fibroblasts and mouse skin. Aging Cell 19, e13072 (2020).
Zhao, J. et al. ATM is a key driver of NF-κB-dependent DNA-damage-induced senescence, stem cell dysfunction and aging. Aging 12, 4688–4710 (2020).
Cockayne, E. A. Dwarfism with retinal atrophy and deafness. Arch. Dis. Child. 11, 1–8 (1936).
Karikkineth, A. C., Scheibye-Knudsen, M., Fivenson, E., Croteau, D. L. & Bohr, V. A. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res. Rev. 33, 3–17 (2017).
Laugel, V. Cockayne syndrome: the expanding clinical and mutational spectrum. Mech. Ageing Dev. 134, 161–170 (2013).
Majora, M. et al. HDAC inhibition improves autophagic and lysosomal function to prevent loss of subcutaneous fat in a mouse model of Cockayne syndrome. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aam7510 (2018).
Kajitani, G. S. et al. Neurovascular dysfunction and neuroinflammation in a Cockayne syndrome mouse model. Aging 13, 22710–22731 (2021).
Berquist, B. R., Canugovi, C., Sykora, P., Wilson, D. M. III & Bohr, V. A. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 40, 8392–8405 (2012).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Chung, K. W. et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 30, 784–799.e5 (2019).
Oshima, J., Sidorova, J. M. & Monnat, R. J. Jr Werner syndrome: clinical features, pathogenesis and potential therapeutic interventions. Ageing Res. Rev. 33, 105–114 (2017).
Turaga, R. V. et al. The Werner syndrome protein affects the expression of genes involved in adipogenesis and inflammation in addition to cell cycle and DNA damage responses. Cell Cycle 8, 2080–2092 (2009).
Davis, T. & Kipling, D. Werner syndrome as an example of inflamm-aging: possible therapeutic opportunities for a progeroid syndrome? Rejuvenation Res. 9, 402–407 (2006).
Goto, M. et al. Multiplex cytokine analysis of Werner syndrome. Intractable Rare Dis. Res. 4, 190–197 (2015).
Goto, M., Chiba, J., Matsuura, M., Iwaki-Egawa, S. & Watanabe, Y. Inflammageing assessed by MMP9 in normal Japanese individuals and the patients with Werner syndrome. Intractable Rare Dis. Res. 5, 103–108 (2016).
Goto, M. et al. Aging-associated inflammation in healthy Japanese individuals and patients with Werner syndrome. Exp. Gerontol. 47, 936–939 (2012).
Yu, Q. et al. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep. 11, 785–797 (2015).
Freund, A., Patil, C. K. & Campisi, J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30, 1536–1548 (2011).
Amor-Gueret, M. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett. 236, 1–12 (2006).
Rao, V. A. et al. Endogenous γ-H2AX–ATM–Chk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 5, 713–724 (2007).
Gratia, M. et al. Bloom syndrome protein restrains innate immune sensing of micronuclei by cGAS. J. Exp. Med. 216, 1199–1213 (2019).
Rehman, R. et al. Noncanonical role for Ku70/80 in the prevention of allergic airway inflammation via maintenance of airway epithelial cell organelle homeostasis. Am. J. Physiol. Lung Cell Mol. Physiol. 319, L728–L741 (2020).
Sun, X. et al. DNA-PK deficiency potentiates cGAS-mediated antiviral innate immunity. Nat. Commun. 11, 6182 (2020).
Ferguson, B. J., Mansur, D. S., Peters, N. E., Ren, H. & Smith, G. L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 1, e00047 (2012).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: the Trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Sirbu, B. M. & Cortez, D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 5, a012724 (2013).
Boder, E. & Sedgwick, R. P. Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics 21, 526–554 (1958).
Shiloh, Y. & Lederman, H. M. Ataxia-telangiectasia (A-T): an emerging dimension of premature ageing. Ageing Res. Rev. 33, 76–88 (2017).
Zaki-Dizaji, M., Akrami, S. M., Azizi, G., Abolhassani, H. & Aghamohammadi, A. Inflammation, a significant player of ataxia-telangiectasia pathogenesis? Inflamm. Res. 67, 559–570 (2018).
McGrath-Morrow, S. A. et al. Elevated serum IL-8 levels in ataxia telangiectasia. J. Pediatr. 156, 682–684 e681 (2010).
McGrath-Morrow, S. A., Collaco, J. M., Detrick, B. & Lederman, H. M. Serum interleukin-6 levels and pulmonary function in ataxia-telangiectasia. J. Pediatr. 171, 256–261.e1 (2016).
Saunders, R. A. et al. Elevated inflammatory responses and targeted therapeutic intervention in a preclinical mouse model of ataxia-telangiectasia lung disease. Sci. Rep. 11, 4268 (2021).
Gul, E. et al. Type I IFN-related NETosis in ataxia telangiectasia and Artemis deficiency. J. Allergy Clin. Immunol. 142, 246–257 (2018).
Song, X., Ma, F. & Herrup, K. Accumulation of cytoplasmic DNA due to ATM deficiency activates the microglial viral response system with neurotoxic consequences. J. Neurosci. 39, 6378–6394 (2019).
Eriksson, M. et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423, 293–298 (2003).
Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. & Spann, T. P. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16, 533–547 (2002).
Goldman, R. D. et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. Usa. 101, 8963–8968 (2004).
Gonzalo, S. & Kreienkamp, R. DNA repair defects and genome instability in Hutchinson–Gilford progeria syndrome. Curr. Opin. Cell Biol. 34, 75–83 (2015).
Gonzalez-Dominguez, A. et al. Inhibition of the NLRP3 inflammasome improves lifespan in animal murine model of Hutchinson–Gilford progeria. EMBO Mol. Med. 13, e14012 (2021).
Mu, X. et al. Cytoskeleton stiffness regulates cellular senescence and innate immune response in Hutchinson–Gilford progeria syndrome. Aging Cell 19, e13152 (2020).
Osorio, F. G. et al. Nuclear lamina defects cause ATM-dependent NF-κB activation and link accelerated aging to a systemic inflammatory response. Genes Dev. 26, 2311–2324 (2012).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Mouse Genome Sequencing, C. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Dewannieux, M. & Heidmann, T. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 3, 646–656 (2013).
Hancks, D. C. & Kazazian, H. H. Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203 (2012).
Richardson, S. R. et al. The Influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MDNA3-0061-2014 (2015).
Bourque, G. et al. Ten things you should know about transposable elements. Genome Biol. 19, 199 (2018).
Deininger, P. L. & Batzer, M. A. Alu repeats and human disease. Mol. Genet. Metab. 67, 183–193 (1999).
Kury, P. et al. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 24, 379–394 (2018).
Boissinot, S. & Sookdeo, A. The evolution of LINE-1 in vertebrates. Genome Biol. Evol. 8, 3485–3507 (2016).
Gorbunova, V. et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53 (2021).
Di Giacomo, M. et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol. Cell 50, 601–608 (2013).
Yang, F. & Wang, P. J. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin. Cell Dev. Biol. 59, 118–125 (2016).
Kazazian, H. H. Jr & Moran, J. V. Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017).
Rodic, N. LINE-1 activity and regulation in cancer. Front. Biosci. 23, 1680–1686 (2018).
Burns, K. H. Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424 (2017).
Belancio, V. P., Roy-Engel, A. M. & Deininger, P. L. All y’all need to know ‘bout retroelements in cancer. Semin. Cancer Biol. 20, 200–210 (2010).
Rodic, N. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286 (2014).
Sciamanna, I., De Luca, C. & Spadafora, C. The reverse transcriptase encoded by LINE-1 retrotransposons in the genesis, progression, and therapy of cancer. Front. Chem. 4, 6 (2016).
Burns, K. H. Our conflict with transposable elements and its implications for human disease. Annu. Rev. Pathol. 15, 51–70 (2020).
Thomas, C. A. et al. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21, 319–331.e8 (2017).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019). This paper demonstrates that LINE1 elements induce inflammation in senescent cells via activation of cGAS–STING.
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). This paper links LINE1 elements to activation of autoimmunity.
Simon, M. et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885.e5 (2019). This paper demonstrates that activation of LINE1 elements in aged and SIRT6 knockout mice triggers inflammation via formation of cytoplasmic DNA and activation of cGAS–STING.
Tiwari, B. et al. p53 directly represses human LINE1 transposons. Genes Dev. 34, 1439–1451 (2020).
Bregnard, C. et al. Upregulated LINE-1 activity in the fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine 8, 184–194 (2016).
Tunbak, H. et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat. Commun. 11, 5387 (2020).
Kaneko, H. et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 (2011).
Tao, S. S. et al. TREX1 as a potential therapeutic target for autoimmune and inflammatory diseases. Curr. Pharm. Des. 25, 3239–3247 (2019).
Fukuda, S. et al. Alu complementary DNA is enriched in atrophic macular degeneration and triggers retinal pigmented epithelium toxicity via cytosolic innate immunity. Sci. Adv. 7, eabj3658 (2021).
Morita, M. et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell Biol. 24, 6719–6727 (2004).
Ahmad, S. et al. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810.e13 (2018).
Fukuda, S. et al. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2022751118 (2021).
Miller, K. N. et al. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184, 5506–5526 (2021).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Van Meter, M. et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5, 5011 (2014).
Teo, Y. V. et al. Cell-free DNA as a biomarker of aging. Aging Cell 18, e12890 (2019).
Mahmood, W. et al. Aging-associated distinctive DNA methylation changes of LINE-1 retrotransposons in pure cell-free DNA from human blood. Sci. Rep. 10, 22127 (2020).
Ponomaryova, A. A. et al. Aberrant methylation of LINE-1 transposable elements: a search for cancer biomarkers. Cells https://doi.org/10.3390/cells9092017 (2020).
Schmid, C. W. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 19, 5613–5617 (1991).
Gentilini, D. et al. Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age 35, 1961–1973 (2013).
Jintaridth, P. & Mutirangura, A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol. Genomics 41, 194–200 (2010).
Franceschi, C. et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 908, 244–254 (2000).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). Together with reference 120, this work demonstrates the tumour-suppressive roles by inhibiting DNA methylation.
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563.e19 (2018).
Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019).
Mehdipour, P. et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020).
Gu, Z. et al. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia. Nat. Genet. 53, 672–682 (2021). This study demonstrates that LINE1 has a tumour-suppressive function in myeloid leukaemia.
Zhao, Y. et al. Transposon-triggered innate immune response confers cancer resistance to the blind mole rat. Nat. Immunol. 22, 1219–1230 (2021). This study reports that RTEs can naturally serve as tumour suppressors through activation of innate immune response.
Gorbunova, V. et al. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl Acad. Sci. USA 109, 19392–19396 (2012).
Campisi, J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522 (2005).
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961).
Campisi, J. From cells to organisms: can we learn about aging from cells in culture? Exp. Gerontol. 36, 607–618 (2001).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010).
Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). This study explains the source of SASP, linking the inflammation to cytoplasmic nucleic acid sensing in senescent cells.
Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 202, 129–143 (2013).
Freund, A., Laberge, R. M., Demaria, M. & Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075 (2012).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Vizioli, M. G. et al. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 34, 428–445 (2020).
Miller, K. N., Dasgupta, N., Liu, T., Adams, P. D. & Vizioli, M. G. Cytoplasmic chromatin fragments-from mechanisms to therapeutic potential. eLife https://doi.org/10.7554/eLife.63728 (2021).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. Usa. 114, E4612–E4620 (2017). This study demonstrates the essential role of cGAS in cellular senescence.
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Honda, K., Takaoka, A. & Taniguchi, T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017). This paper establishes the role of cGAS as a driver of senescence and inflammation via the detection of cytoplasmic DNA fragments.
Chen, Y. et al. p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J. Exp. Med. 214, 991–1010 (2017).
Aarreberg, L. D. et al. Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS–STING. Mol. Cell 74, 801–815.e6 (2019).
Zhao, B. et al. Topoisomerase 1 cleavage complex enables pattern recognition and inflammation during senescence. Nat. Commun. 11, 908 (2020).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019).
Kranzusch, P. J. et al. Ancient origin of cGAS–STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015).
Evans, C. J. & Aguilera, R. J. DNase II: genes, enzymes and function. Gene 322, 1–15 (2003).
Rodero, M. P. et al. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8, 2176 (2017).
Lan, Y. Y. et al. Extranuclear DNA accumulates in aged cells and contributes to senescence and inflammation. Aging Cell 18, e12901 (2019).
Jutte, B. B. et al. Intercellular cGAMP transmission induces innate immune activation and tissue inflammation in Trex1 deficiency. iScience 24, 102833 (2021).
Berndt, N. et al. Photosensitivity and cGAS-dependent type I IFN activation in lupus patients with TREX1 deficiency. J. Invest. Dermatol. https://doi.org/10.1016/j.jid.2021.04.037 (2021).
Takahashi, A. et al. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 9, 1249 (2018).
Han, X. et al. Autolysosomal degradation of cytosolic chromatin fragments antagonizes oxidative stress-induced senescence. J. Biol. Chem. 295, 4451–4463 (2020).
Niehrs, C. & Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 21, 167–178 (2020).
Yu, K., Chedin, F., Hsieh, C. L., Wilson, T. E. & Lieber, M. R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451 (2003).
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E. & Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007).
Sanz, L. A. et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016).
Cheng, L., Wang, W., Yao, Y. & Sun, Q. Mitochondrial RNase H1 activity regulates R-loop homeostasis to maintain genome integrity and enable early embryogenesis in Arabidopsis. PLoS Biol. 19, e3001357 (2021).
Cerritelli, S. M. & Crouch, R. J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276, 1494–1505 (2009).
Skourti-Stathaki, K., Proudfoot, N. J. & Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011).
Chang, E. Y. et al. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J. Cell Biol. 216, 3991–4005 (2017).
Marabitti, V. et al. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 47, 3485–3502 (2019).
D’Alessandro, G. et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 9, 5376 (2018).
Kennedy, R. D. & D’Andrea, A. D. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940 (2005).
Bhatia, V. et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014).
Hatchi, E. et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015).
Sollier, J. et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014).
Gan, W. et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011).
Costantino, L. & Koshland, D. Genome-wide map of R-loop-induced damage reveals how a subset of R-loops contributes to genomic instability. Mol. Cell 71, 487–497 e483 (2018).
Goulielmaki, E. et al. The splicing factor XAB2 interacts with ERCC1–XPF and XPG for R-loop processing. Nat. Commun. 12, 3153 (2021).
Cristini, A. et al. Dual processing of R-loops and topoisomerase i induces transcription-dependent DNA double-strand breaks. Cell Rep. 28, 3167–3181.e6 (2019).
Promonet, A. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11, 3940 (2020).
Tian, X. et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622–638 e622 (2019). This paper establishes a link between more efficient DSB repair and longer lifespan via the activity of SIRT6 protein.
Niedernhofer, L. J. et al. Nuclear genomic instability and aging. Annu. Rev. Biochem. 87, 295–322 (2018).
Grunseich, C. et al. Senataxin mutation reveals how R-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell 69, 426–437.e7 (2018).
Crow, Y. J. et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 (2006).
Suraweera, A. et al. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J. Cell Biol. 177, 969–979 (2007).
Becherel, O. J. et al. A new model to study neurodegeneration in ataxia oculomotor apraxia type 2. Hum. Mol. Genet. 24, 5759–5774 (2015).
Gunther, C. et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 125, 413–424 (2015).
Barnhoorn, S. et al. Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG deficiency. PLoS Genet. 10, e1004686 (2014).
Trego, K. S. et al. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle 10, 1998–2007 (2011).
Zeller, P. et al. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48, 1385–1395 (2016).
Padeken, J. et al. Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev. 33, 436–451 (2019).
Li, C. L. et al. Region-specific H3K9me3 gain in aged somatic tissues in Caenorhabditis elegans. PLoS Genet. 17, e1009432 (2021).
Weinreb, J. T. et al. Excessive R-loops trigger an inflammatory cascade leading to increased HSPC production. Dev. Cell 56, 627–640.e5 (2021).
Chatzidoukaki, O. et al. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci. Adv. 7, eabj5769 (2021).
Chen, H. et al. cGAS suppresses genomic instability as a decelerator of replication forks. Sci. Adv. https://doi.org/10.1126/sciadv.abb8941 (2020).
Yang, J.-H. et al. Erosion of the epigenetic landscape and loss of cellular identity as a cause of aging in mammals. Preprint at bioRxiv https://doi.org/10.1101/808642 (2019).
Hayano, M. et al. DNA break-induced epigenetic drift as a cause of mammalian aging. Preprint at bioRxiv https://doi.org/10.1101/808659 (2019).
Kim, J., Sturgill, D., Tran, A. D., Sinclair, D. A. & Oberdoerffer, P. Controlled DNA double-strand break induction in mice reveals post-damage transcriptome stability. Nucleic Acids Res. 44, e64 (2016).
Sedivy, J. M., Banumathy, G. & Adams, P. D. Aging by epigenetics — a consequence of chromatin damage? Exp. Cell Res. 314, 1909–1917 (2008).
Tsurumi, A. & Li, W. X. Global heterochromatin loss: a unifying theory of aging? Epigenetics 7, 680–688 (2012).
Vaquero, A. et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16, 93–105 (2004).
Vaquero, A. et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450, 440–444 (2007).
Oberdoerffer, P. et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 (2008).
Mao, Z. et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 (2011).
Haag, S. M. et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018).
Lama, L. et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 10, 2261 (2019).
Dai, J. et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176, 1447–1460.e14 (2019).
Piscianz, E. et al. Reappraisal of antimalarials in interferonopathies: new perspectives for old drugs. Curr. Med. Chem. 25, 2797–2810 (2018).
Broz, P. & Dixit, V. M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016).
Coll, R. C. et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015).
Fowler, B. J. et al. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science 346, 1000–1003 (2014).
Yuan, S., Shi, Y., Guo, K. & Tang, S. J. Nucleoside reverse transcriptase inhibitors (NRTIs) induce pathological pain through Wnt5a-mediated neuroinflammation in aging mice. J. Neuroimmune Pharmacol. 13, 230–236 (2018).
Dai, L., Huang, Q. & Boeke, J. D. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 12, 18 (2011).
Petersen, A. M. & Pedersen, B. K. The anti-inflammatory effect of exercise. J. Appl. Physiol. 98, 1154–1162 (2005).
Ott, B. et al. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci. Rep. 7, 11955 (2017).
Meydani, S. N. et al. Long-term moderate calorie restriction inhibits inflammation without impairing cell-mediated immunity: a randomized controlled trial in non-obese humans. Aging 8, 1416–1431 (2016).
Gabande-Rodriguez, E., Gomez de Las Heras, M. M. & Mittelbrunn, M. Control of inflammation by calorie restriction mimetics: on the crossroad of autophagy and mitochondria. Cells https://doi.org/10.3390/cells9010082 (2019).
Wongrakpanich, S., Wongrakpanich, A., Melhado, K. & Rangaswami, J. A comprehensive review of non-steroidal anti-inflammatory drug use in the elderly. Aging Dis. 9, 143–150 (2018).
Mao, Z. et al. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc. Natl Acad. Sci. USA 109, 11800–11805 (2012).
Huang, J. et al. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS ONE 3, e1710 (2008).
Chen, Y. et al. A PARP1–BRG1–SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites. Nucleic Acids Res. 47, 8563–8580 (2019).
Ocampo, A. et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733.e12 (2016).
Sarkar, T. J. et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 11, 1545 (2020).
Klein, M. A. et al. Mechanism of activation for the sirtuin 6 protein deacylase. J. Biol. Chem. 295, 1385–1399 (2020).
Huang, Z. et al. Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 14, 1118–1126 (2018).
Shang, J. L., Ning, S. B., Chen, Y. Y., Chen, T. X. & Zhang, J. MDL-800, an allosteric activator of SIRT6, suppresses proliferation and enhances EGFR-TKIs therapy in non-small cell lung cancer. Acta Pharmacol. Sin. 42, 120–131 (2021).
Chen, Y. et al. The SIRT6 activator MDL-800 improves genomic stability and pluripotency of old murine-derived iPS cells. Aging Cell 19, e13185 (2020).
Bonkowski, M. S. & Sinclair, D. A. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 (2016).
Rahnasto-Rilla, M. K. et al. The identification of a SIRT6 activator from brown algae Fucus distichus. Mar. Drugs https://doi.org/10.3390/md15060190 (2017).
Price, B. D. & D’Andrea, A. D. Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 (2013).
Hauer, M. H. & Gasser, S. M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 31, 2204–2221 (2017).
Zhang, W., Qu, J., Liu, G. H. & Belmonte, J. C. I. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 21, 137–150 (2020).
Saul, D. & Kosinsky, R. L. Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22010401 (2021).
Benayoun, B. A., Pollina, E. A. & Brunet, A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 16, 593–610 (2015).
Benayoun, B. A. et al. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709 (2019).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
V.G. is a consultant for Elysium, Centaura, Genflow Bio and Do Not Age. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks P. McHugh, M. Ratnaweera and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Nucleotide excision repair
-
(NER). A DNA repair pathway that recognizes and removes bulky DNA lesions such as those formed by UV light. NER involves damage recognition, nicks flanking the lesion and removal of the damaged DNA strand, filling in the gap by DNA polymerase and ligation of the nicks to restore the intact DNA molecule. Deficiency in NER is associated with cancer predisposition and premature ageing syndromes.
- Senescence-associated secretory phenotype
-
(SASP). A phenotype associated with cellular senescence, which expresses and secretes a wide variety of inflammatory factors including cytokines, chemokines and growth factors.
- DNA double-strand break (DSB) repair
-
A DNA repair pathway that repairs lesions where both DNA strands are broken. There are two major pathways of DSB repair; homologous recombination uses sister chromatid as a template for repair and non-homologous DNA end joining ligates the broken ends without regard for homology. Defects in DNA repair are associated with several premature ageing and cancer predisposition syndromes.
- Inflammageing
-
Chronic, low-grade sterile inflammation frequently observed during ageing.
- Retrotransposons
-
(RTEs). Genetic elements abundant in mammalian genomes that move throughout the genome by a copy–paste mechanism involving reverse transcription and may accumulate in the cytoplasm, triggering inflammation.
- Sterile inflammation
-
Inflammation arising in the absence of a pathogen or external inflammatory stimuli.
- Blind mole rat
-
A subterranean rodent of the Muroidea superfamily, characterized by long maximum lifespan (>21 years) and resistance to cancer.
- HRASV12
-
An oncoprotein of small GTPase HRAS carrying a constitutively activated mutation on codon Val-12.
Rights and permissions
About this article
Cite this article
Zhao, Y., Simon, M., Seluanov, A. et al. DNA damage and repair in age-related inflammation. Nat Rev Immunol 23, 75–89 (2023). https://doi.org/10.1038/s41577-022-00751-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-022-00751-y
This article is cited by
-
Sirtuins in intervertebral disc degeneration: current understanding
Molecular Medicine (2024)
-
Cytosolic DNA sensors in neurodegenerative diseases: from physiological defenders to pathological culprits
EMBO Molecular Medicine (2024)
-
Replicative senescence and high glucose induce the accrual of self-derived cytosolic nucleic acids in human endothelial cells
Cell Death Discovery (2024)
-
Roles and regulation of tRNA-derived small RNAs in animals
Nature Reviews Molecular Cell Biology (2024)
-
Association of food insecurity with successful aging among older Indians: study based on LASI
European Journal of Nutrition (2024)