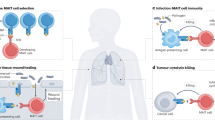
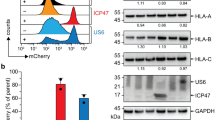
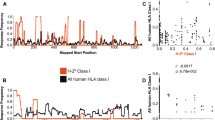
Abstract
Antigen processing and presentation are the cornerstones of adaptive immunity. B cells cannot generate high-affinity antibodies without T cell help. CD4+ T cells, which provide such help, use antigen-specific receptors that recognize major histocompatibility complex (MHC) molecules in complex with peptide cargo. Similarly, eradication of virus-infected cells often depends on cytotoxic CD8+ T cells, which rely on the recognition of peptide–MHC complexes for their action. The two major classes of glycoproteins entrusted with antigen presentation are the MHC class I and class II molecules, which present antigenic peptides to CD8+ T cells and CD4+ T cells, respectively. This Review describes the essentials of antigen processing and presentation. These pathways are divided into six discrete steps that allow a comparison of the various means by which antigens destined for presentation are acquired and how the source proteins for these antigens are tagged for degradation, destroyed and ultimately displayed as peptides in complex with MHC molecules for T cell recognition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Vyas, J. M., Van der Veen, A. G. & Ploegh, H. L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8, 607–618 (2008).
Blees, A. et al. Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 (2017).
Trowitzsch, S. & Tampe, R. Multifunctional chaperone and quality control complexes in adaptive immunity. Annu. Rev. Biophys. 49, 135–161 (2020).
Jensen, P. E. Recent advances in antigen processing and presentation. Nat. Immunol. 8, 1041–1048 (2007).
Call, M. E. & Wucherpfennig, K. W. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 23, 101–125 (2005).
Martin, F. & Chan, A. C. B cell immunobiology in disease: evolving concepts from the clinic. Annu. Rev. Immunol. 24, 467–496 (2006).
Lizee, G. et al. Control of dendritic cell cross-presentation by the major histocompatibility complex class I cytoplasmic domain. Nat. Immunol. 4, 1065–1073 (2003).
Reeves, E. & James, E. Antigen processing and immune regulation in the response to tumours. Immunology 150, 16–24 (2017).
Fernando, M. M. et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 4, e1000024 (2008).
Neefjes, J. et al. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011).
Blum, J. S., Wearsch, P. A. & Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013).
Cresswell, P. et al. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol. Rev. 207, 145–157 (2005).
van Kasteren, S. I. et al. Chemical biology of antigen presentation by MHC molecules. Curr. Opin. Immunol. 26, 21–31 (2014).
Kyewski, B. & Klein, L. A central role for central tolerance. Annu. Rev. Immunol. 24, 571–606 (2006).
Lee, J. W. et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat. Immunol. 8, 181–190 (2007).
Kyewski, B. & Derbinski, J. Self-representation in the thymus: an extended view. Nat. Rev. Immunol. 4, 688–698 (2004).
Mayassi, T. et al. A multilayered immune system through the lens of unconventional T cells. Nature 595, 501–510 (2021).
Adams, E. J. & Luoma, A. M. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31, 529–561 (2013).
Stern, L. J. & Wiley, D. C. Antigenic peptide binding by class I and class II histocompatibility proteins. Behring Inst. Mitt. 2, 245–251 (1994).
Tumer, G., Simpson, B. and Roberts, T. K. Genetics, Human Major Histocompatibility Complex (MHC) (StatPearls, 2021)
Choo, S. Y. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med. J. 48, 11–23 (2007).
Shiina, T. et al. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 54, 15–39 (2009).
Shiina, T. et al. Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150, 127–138 (2017).
Matsumura, M. et al. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 257, 927–934 (1992).
Bouvier, M. & Wiley, D. C. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science 265, 398–402 (1994).
Zacharias, M. & Springer, S. Conformational flexibility of the MHC class I α1-α2 domain in peptide bound and free states: a molecular dynamics simulation study. Biophys. J. 87, 2203–2214 (2004).
Van Rhijn, I. et al. Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 15, 643–654 (2015).
Silva, A. P. D. & Gallardo, R. A. The chicken MHC: insights into genetic resistance, immunity, and inflammation following infectious bronchitis virus infections. Vaccines (Basel) 8, 637 (2020).
Miller, M. M. & Taylor, R. L. Jr. Brief review of the chicken major histocompatibility complex: the genes, their distribution on chromosome 16, and their contributions to disease resistance. Poult. Sci. 95, 375–392 (2016).
Chicz, R. M. et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 358, 764–768 (1992).
Abualrous, E. T., Sticht, J. & Freund, C. Major histocompatibility complex (MHC) class I and class II proteins: impact of polymorphism on antigen presentation. Curr. Opin. Immunol. 70, 95–104 (2021).
Flajnik, M. F. & Kasahara, M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 47–59 (2010).
Bontrop, R. E. Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum. Immunol. 67, 388–397 (2006).
Kasahara, M. The chromosomal duplication model of the major histocompatibility complex. Immunol. Rev. 167, 17–32 (1999).
Robinson, J. et al. IPD-IMGT/HLA Database. Nucleic Acids Res. 48, D948–D955 (2020).
Kelly, A. & Trowsdale, J. Genetics of antigen processing and presentation. Immunogenetics 71, 161–170 (2019).
Tomasec, P. et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000).
Larsen, M. H. & Hviid, T. V. Human leukocyte antigen-G polymorphism in relation to expression, function, and disease. Hum. Immunol. 70, 1026–1034 (2009).
Wu, H. L. et al. The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J. Immunol. 200, 49–60 (2018).
Dulberger, C. L. et al. Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity 46, 1018–1029 e7 (2017).
Creech, A. L. et al. The role of mass spectrometry and proteogenomics in the advancement of HLA epitope prediction. Proteomics 18, e1700259 (2018).
Thomas, C. & Tampe, R. MHC I chaperone complexes shaping immunity. Curr. Opin. Immunol. 58, 9–15 (2019).
Wieczorek, M. et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 8, 292 (2017).
Truong, H. V. & Sgourakis, N. G. Dynamics of MHC-I molecules in the antigen processing and presentation pathway. Curr. Opin. Immunol. 70, 122–128 (2021).
Zaitoua, A. J., Kaur, A. & Raghavan, M. Variations in MHC class I antigen presentation and immunopeptidome selection pathways. F1000Res. https://doi.org/10.12688/f1000research.26935.1 (2020).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Jiang, J., Natarajan, K. & Margules, D. H. MHC molecules, T cell receptors, natural killer cell receptors, and viral immunoevasins-key elements of adaptive and innate immunity. Adv. Exp. Med. Biol. 1172, 21–62 (2019).
Cosgrove, D. et al. Mice lacking MHC class II molecules. Cell 66, 1051–1066 (1991).
Koller, B. H. et al. Normal development of mice deficient in β2M, MHC class I proteins, and CD8+ T cells. Science 248, 1227–1230 (1990).
Stuart, L. M. & Ezekowitz, R. A. Phagocytosis: elegant complexity. Immunity 22, 539–550 (2005).
Tse, S. M. et al. Differential role of actin, clathrin, and dynamin in Fc gamma receptor-mediated endocytosis and phagocytosis. J. Biol. Chem. 278, 3331–3338 (2003).
Siemasko, K. et al. Cutting edge: signals from the B lymphocyte antigen receptor regulate MHC class II containing late endosomes. J. Immunol. 160, 5203–5208 (1998).
Lankar, D. et al. Dynamics of major histocompatibility complex class II compartments during B cell receptor-mediated cell activation. J. Exp. Med. 195, 461–472 (2002).
Lim, J. P. & Gleeson, P. A. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 89, 836–843 (2011).
Schmid, D., Pypaert, M. & Munz, C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26, 79–92 (2007).
Trombetta, E. S. & Mellman, I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23, 975–1028 (2005).
Wilson, N. S., El-Sukkari, D. & Villadangos, J. A. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood 103, 2187–2195 (2004).
Chancellor, A., Gadola, S. D. & Mansour, S. The versatility of the CD1 lipid antigen presentation pathway. Immunology 154, 196–203 (2018).
Koch, M. et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 6, 819–826 (2005).
Wu, D., Fujio, M. & Wong, C. H. Glycolipids as immunostimulating agents. Bioorg. Med. Chem. 16, 1073–1083 (2008).
Rock, K. L. & Goldberg, A. L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17, 739–779 (1999).
Hewitt, E. W. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110, 163–169 (2003).
Hill, A. & Ploegh, H. Getting the inside out: the transporter associated with antigen processing (TAP) and the presentation of viral antigen. Proc. Natl Acad. Sci. USA 92, 341–343 (1995).
Cruz, F. M. et al. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu. Rev. Immunol. 35, 149–176 (2017).
Hughes, E. A., Hammond, C. & Cresswell, P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl Acad. Sci. USA 94, 1896–1901 (1997).
van Hall, T. et al. The other Janus face of Qa-1 and HLA-E: diverse peptide repertoires in times of stress. Microbes Infect. 12, 910–918 (2010).
Delamarre, L. et al. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307, 1630–1634 (2005).
West, L. C. & Cresswell, P. Expanding roles for GILT in immunity. Curr. Opin. Immunol. 25, 103–108 (2013).
Li, M. et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333, 53–58 (2011).
Yewdell, J. W. & Hickman, H. D. New lane in the information highway: alternative reading frame peptides elicit T cells with potent antiretrovirus activity. J. Exp. Med. 204, 2501–2504 (2007).
Berglund, P. et al. Viral alteration of cellular translational machinery increases defective ribosomal products. J. Virol. 81, 7220–7229 (2007).
Netzer, N. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009).
Dolan, B. P. et al. Distinct pathways generate peptides from defective ribosomal products for CD8+T cell immunosurveillance. J. Immunol. 186, 2065–2072 (2011).
Schubert, U. et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000).
Reits, E. A. et al. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404, 774–778 (2000).
Welchman, R. L., Gordon, C. & Mayer, R. J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 (2005).
Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87 (2005).
Vigneron, N. et al. Peptide splicing by the proteasome. J. Biol. Chem. 292, 21170–21179 (2017).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Bard, J. A. M. et al. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018).
Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017).
Tanaka, K., Mizushima, T. & Saeki, Y. The proteasome: molecular machinery and pathophysiological roles. Biol. Chem. 393, 217–234 (2012).
Hanada, K., Yewdell, J. W. & Yang, J. C. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004).
Vigneron, N. et al. An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004).
Murata, S. et al. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat. Immunol. 19, 923–931 (2018).
Tanaka, K. & Kasahara, M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol. Rev. 163, 161–176 (1998).
Hattori, A. & Tsujimoto, M. Endoplasmic reticulum aminopeptidases: biochemistry, physiology and pathology. J. Biochem. 154, 219–228 (2013).
Suzuki, T., Huang, C. & Fujihira, H. The cytoplasmic peptide:N-glycanase (NGLY1) - structure, expression and cellular functions. Gene 577, 1–7 (2016).
Rodgers, J. R. & Cook, R. G. MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol. 5, 459–471 (2005).
McWilliam, H. E. G. et al. Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl Acad. Sci. USA 117, 24974–24985 (2020).
Villadangos, J. A. et al. Proteases involved in MHC class II antigen presentation. Immunol. Rev. 172, 109–120 (1999).
Honey, K. & Rudensky, A. Y. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3, 472–482 (2003).
Wang, Y. et al. How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes. Sci. Immunol. 4, eaav7517 (2019).
Winchester, B. Lysosomal metabolism of glycoproteins. Glycobiology 15, 1R–15R (2005).
Parcej, D. & Tampe, R. ABC proteins in antigen translocation and viral inhibition. Nat. Chem. Biol. 6, 572–580 (2010).
Eggensperger, S. & Tampe, R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol. Chem. 396, 1059–1072 (2015).
Thomas, C. & Tampe, R. Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636 (2020).
Grossmann, N. et al. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat. Commun. 5, 5419 (2014).
Gubler, B. et al. Substrate selection by transporters associated with antigen processing occurs during peptide binding to TAP. Mol. Immunol. 35, 427–433 (1998).
Uebel, S. et al. Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc. Natl Acad. Sci. USA 94, 8976–8981 (1997).
Serwold, T. et al. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 (2002).
Saric, T. et al. An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 (2002).
York, I. A. et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 3, 1177–1184 (2002).
Roche, P. A. & Cresswell, P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature 345, 615–618 (1990).
Reich, M. et al. Invariant chain processing is independent of cathepsin variation between primary human B cells/dendritic cells and B-lymphoblastoid cells. Cell Immunol. 269, 96–103 (2011).
Williams, D. B. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 119, 615–623 (2006).
Raghavan, M. et al. MHC class I assembly: out and about. Trends Immunol. 29, 436–443 (2008).
Garbi, N. et al. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat. Immunol. 1, 234–238 (2000).
Busch, R. et al. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol. Rev. 207, 242–260 (2005).
Poluektov, Y. O., Kim, A. & Sadegh-Nasseri, S. HLA-DO and its role in MHC class II antigen presentation. Front. Immunol. 4, 260 (2013).
Sollid, L. M., Pos, W. & Wucherpfennig, K. W. Molecular mechanisms for contribution of MHC molecules to autoimmune diseases. Curr. Opin. Immunol. 31, 24–30 (2014).
Klein, L. et al. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844 (2009).
Baker, B. M. et al. Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism. Immunol. Rev. 250, 10–31 (2012).
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
ten Broeke, T., Wubbolts, R. & Stoorvogel, W. MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harb. Perspect. Biol. 5, a016873 (2013).
Watts, C., West, M. A. & Zaru, R. TLR signalling regulated antigen presentation in dendritic cells. Curr. Opin. Immunol. 22, 124–130 (2010).
Bhati, M. et al. The versatility of the αβ T-cell antigen receptor. Protein Sci. 23, 260–272 (2014).
Mittal, S. K. & Roche, P. A. Suppression of antigen presentation by IL-10. Curr. Opin. Immunol. 34, 22–27 (2015).
Paul, P. et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell 145, 268–283 (2011).
van de Weijer, M. L., Luteijn, R. D. & Wiertz, E. J. Viral immune evasion: lessons in MHC class I antigen presentation. Semin. Immunol. 27, 125–137 (2015).
Loureiro, J. & Ploegh, H. L. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv. Immunol. 92, 225–305 (2006).
Bauer, D. & Tampe, R. Herpes viral proteins blocking the transporter associated with antigen processing TAP — from genes to function and structure. Curr. Top. Microbiol. Immunol. 269, 87–99 (2002).
Berry, R. et al. Modulation of innate and adaptive immunity by cytomegaloviruses. Nat. Rev. Immunol. 20, 113–127 (2020).
Lin, J. et al. A negative feedback modulator of antigen processing evolved from a frameshift in the cowpox virus genome. PLoS Pathog. 10, e1004554 (2014).
Browne, H. et al. A complex between the MHC class I homologue encoded by human cytomegalovirus and β2 microglobulin. Nature 347, 770–772 (1990).
Farrell, H. E. et al. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 386, 510–514 (1997).
Dhatchinamoorthy, K., Colbert, J. D. & Rock, K. L. Cancer immune evasion through loss of MHC class I antigen presentation. Front. Immunol. 12, 636568 (2021).
Hong, M., Clubb, J. D. & Chen, Y. Y. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell 38, 473–488 (2020).
Kincaid, E. Z. et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 13, 129–135 (2011).
Yan, J. et al. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J. Exp. Med. 203, 647–659 (2006).
Miller, Z. et al. Inhibitors of the immunoproteasome: current status and future directions. Curr. Pharm. Des. 19, 4140–4151 (2013).
Rock, K. L. et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 (1994).
Van Kaer, L. et al. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4−8+ T cells. Cell 71, 1205–1214 (1992).
Colbert, J. D., Cruz, F. M. & Rock, K. L. Cross-presentation of exogenous antigens on MHC I molecules. Curr. Opin. Immunol. 64, 1–8 (2020).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Parekh, V. V. et al. Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8α+ dendritic cells. Proc. Natl Acad. Sci. USA 114, E6371–E6380 (2017).
Mintern, J. D. et al. Differential use of autophagy by primary dendritic cells specialized in cross-presentation. Autophagy 11, 906–917 (2015).
Blander, J. M. Regulation of the cell biology of antigen cross-presentation. Annu. Rev. Immunol. 36, 717–753 (2018).
Blander, J. M. The comings and goings of MHC class I molecules herald a new dawn in cross-presentation. Immunol. Rev. 272, 65–79 (2016).
Theisen, D. J. et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018).
Cebrian, I. et al. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 (2011).
Barbet, G. et al. TAP dysfunction in dendritic cells enables noncanonical cross-presentation for T cell priming. Nat. Immunol. 22, 497–509 (2021).
Nair-Gupta, P. et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell 158, 506–521 (2014).
Segura, E. & Amigorena, S. Cross-presentation in mouse and human dendritic cells. Adv. Immunol. 127, 1–31 (2015).
Jongsma, M. L. et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 166, 152–166 (2016).
Kula, T. et al. T-scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178, 1016–1028 e13 (2019).
Woodham, A. W. et al. In vivo detection of antigen-specific CD8+ T cells by immuno-positron emission tomography. Nat. Methods 17, 1025–1032 (2020).
Stopfer, L. E. et al. Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat. Commun. 11, 2760 (2020).
Jhunjhunwala, S., Hammer, C. & Delamarre, L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 21, 298–312 (2021).
Bassani-Sternberg, M. et al. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteom. 14, 658–673 (2015).
Dunn, G. P., Koebel, C. M. & Schreiber, R. D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6, 836–848 (2006).
Bhalla, N., Brooker, R. & Brada, M. Combining immunotherapy and radiotherapy in lung cancer. J. Thorac. Dis. 10, S1447–S1460 (2018).
Janeway, C. A. Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Borbulevych, O. Y., Piepenbrink, K. H. & Baker, B. M. Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J. Immunol. 186, 2950–2958 (2011).
Li, Y. et al. Structural basis for the presentation of tumor-associated MHC class II-restricted phosphopeptides to CD4+ T cells. J. Mol. Biol. 399, 596–603 (2010).
Zajonc, D. M. et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat. Immunol. 6, 810–818 (2005).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
da Fonseca, P. C. & Morris, E. P. Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat. Commun. 6, 7573 (2015).
Fisette, O., Schroder, G. F. & Schafer, L. V. Atomistic structure and dynamics of the human MHC-I peptide-loading complex. Proc. Natl Acad. Sci. USA 117, 20597–20606 (2020).
Pos, W. et al. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell 151, 1557–1568 (2012).
Acknowledgements
The authors thank P. Cresswell and J. Strominger for reading an earlier version of the manuscript. They also thank members of the Ploegh laboratory for their input.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks L. Fugger, R. Tampe and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Pishesha, N., Harmand, T.J. & Ploegh, H.L. A guide to antigen processing and presentation. Nat Rev Immunol 22, 751–764 (2022). https://doi.org/10.1038/s41577-022-00707-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-022-00707-2
This article is cited by
-
Emerging mechanisms of the unfolded protein response in therapeutic resistance: from chemotherapy to Immunotherapy
Cell Communication and Signaling (2024)
-
A new chromosome-scale duck genome shows a major histocompatibility complex with several expanded multigene families
BMC Biology (2024)
-
Engineered EVs with pathogen proteins: promising vaccine alternatives to LNP-mRNA vaccines
Journal of Biomedical Science (2024)
-
A guide to thymic selection of T cells
Nature Reviews Immunology (2024)
-
The emerging family of RORγt+ antigen-presenting cells
Nature Reviews Immunology (2024)