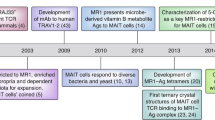
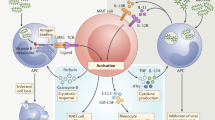
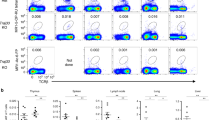
Abstract
MHC antigen presentation plays a fundamental role in adaptive and semi-invariant T cell immunity. Distinct MHC molecules bind antigens that differ in chemical structure, origin and location and present them to specialized T cells. MHC class I-related protein 1 (MR1) presents a range of small molecule antigens to MR1-restricted T (MR1T) lymphocytes. The best studied MR1 ligands are derived from microbial metabolism and are recognized by a major class of MR1T cells known as mucosal-associated invariant T (MAIT) cells. Here, we describe the MR1 antigen presentation pathway: the known types of antigens presented by MR1, the location where MR1–antigen complexes form, the route followed by the complexes to the cell surface, the mechanisms involved in termination of MR1 antigen presentation and the accessory cellular proteins that comprise the MR1 antigen presentation machinery. The current road map of the MR1 antigen presentation pathway reveals potential strategies for therapeutic manipulation of MR1T cell function and provides a foundation for further studies that will lead to a deeper understanding of MR1-mediated immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pishesha, N., Harmand, T. J. & Ploegh, H. L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764 (2022).
Li, D. & Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 6, 291 (2021).
Barral, D. C. & Brenner, M. B. CD1 antigen presentation: how it works. Nat. Rev. Immunol. 7, 929–941 (2007).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010).
Gold, M. C. et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8, e1000407 (2010).
McWilliam, H. E. et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 17, 531–537 (2016). This study outlines the trafficking pathway followed by MR1 from its synthesis in the ER to its degradation in endosomes and the role of VitBAg in regulation of MR1 expression.
Jeffery, H. C. et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J. Hepatol. 64, 1118–1127 (2016).
Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). This study reveals that MR1 binds and presents VitBAgs to MAIT cells.
Corbett, A. J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). This study identifies the most potent MR1 antigens, the 5-A-RU-derived pyrimidines such as 5-OP-RU.
García-Angulo, V. A. Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 43, 196–209 (2017).
Tilloy, F. et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 (1999).
Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003). This study outlines the discovery that MAIT cells are restricted by MR1.
Porcelli, S., Yockey, C. E., Brenner, M. B. & Balk, S. P. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8– α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J. Exp. Med. 178, 1–16 (1993).
Kjer-Nielsen, L. et al. An overview on the identification of MAIT cell antigens. Immunol. Cell Biol. 96, 573–587 (2018).
Gherardin, N. A., McCluskey, J., Rossjohn, J. & Godfrey, D. I. The diverse family of MR1-restricted T cells. J. Immunol. 201, 2862–2871 (2018).
Godfrey, D. I., Koay, H.-F., McCluskey, J. & Gherardin, N. A. The biology and functional importance of MAIT cells. Nat. Immunol. 20, 1110–1128 (2019).
Lantz, O. & Legoux, F. MAIT cells: an historical and evolutionary perspective. Immunol. Cell Biol. 96, 564–572 (2018).
Koay, H.-F. et al. Diverse MR1-restricted T cells in mice and humans. Nat. Commun. 10, 2243 (2019).
Martin, E. et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7, e54 (2009).
Koay, H. F. et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17, 1300–1311 (2016).
Gold, M. C. et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 6, 35–44 (2013).
Legoux, F. et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019). This study shows that MR1 expressed in the thymus presents antigen from peripheral tissues for MAIT cell development.
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019).
Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 (2013).
Gherardin, N. A. et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol. Cell Biol. 96, 507–525 (2018).
Meierovics, A., Yankelevich, W. J. & Cowley, S. C. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl Acad. Sci. USA 110, E3119–E3128 (2013).
Le Bourhis, L. et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 9, e1003681 (2013).
Wang, H. et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9, 3350 (2018).
Leng, T. et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 28, 3077–3091.e5 (2019).
Du Halgouet, A. et al. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity 56, 78–92.e6 (2023).
Smith, A. D. et al. Microbiota of MR1 deficient mice confer resistance against Clostridium difficile infection. PLoS ONE 14, e0223025 (2019).
Varelias, A. et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J. Clin. Invest. 128, 1919–1936 (2018).
Crowther, M. D. & Sewell, A. K. The burgeoning role of MR1-restricted T-cells in infection, cancer and autoimmune disease. Curr. Opin. Immunol. 69, 10–17 (2021).
Shibata, K. et al. Symbiotic bacteria-dependent expansion of MR1-reactive T cells causes autoimmunity in the absence of Bcl11b. Nat. Commun. 13, 6948 (2022).
Gherardin, N. A. et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44, 32–45 (2016).
Vacchini, A., Chancellor, A., Spagnuolo, J., Mori, L. & De Libero, G. MR1-restricted T cells are unprecedented cancer fighters. Front. Immunol. 11, 751 (2020).
Crowther, M. D. et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 21, 178–185 (2020). This study finds an MR1T cell clone that appears to recognize cancer antigens presented by MR1.
Lepore, M. et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. eLife 6, e24476 (2017). This study provides evidence of distinct types of MR1-presented cancer antigens recognized by MR1T cells.
Seneviratna, R. et al. Differential antigenic requirements by diverse MR1-restricted T cells. Immunol. Cell Biol. 100, 112–126 (2022).
Blum, J. S., Wearsch, P. A. & Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013).
Rock, K. L., Reits, E. & Neefjes, J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016).
Villadangos, J. A. & Ploegh, H. L. Proteolysis in MHC class II antigen presentation. Immunity 12, 233–239 (2000).
van de Weijer, M. L., Luteijn, R. D. & Wiertz, E. J. Viral immune evasion: lessons in MHC class I antigen presentation. Semin. Immunol. 27, 125–137 (2015).
Awad, W. et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat. Immunol. 21, 400–411 (2020).
Mak, J. Y. et al. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 8, 14599 (2017).
Tastan, C. et al. Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunol. 11, 1591–1605 (2018).
Mondot, S., Boudinot, P. & Lantz, O. MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics 68, 537–548 (2016).
Schmaler, M. et al. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol. 11, 1060–1070 (2018).
Georgel, P., Radosavljevic, M., Macquin, C. & Bahram, S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol. Immunol. 48, 769–775 (2011).
Howson, L. J. et al. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci. Immunol. 5, eabc9492 (2020). This study examines a patient lacking MAIT cells who had a homozygous mutation in MR1 preventing presentation of VitBAg.
Klebanoff, C. A., Rosenberg, S. A. & Restifo, N. P. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat. Med. 22, 26–36 (2016).
McWilliam, H. E., Birkinshaw, R. W., Villadangos, J. A., McCluskey, J. & Rossjohn, J. MR1 presentation of vitamin B-based metabolite ligands. Curr. Opin. Immunol. 34, 28–34 (2015).
Jin, H. et al. Deaza-modification of MR1 ligands modulates recognition by MR1-restricted T cells. Sci. Rep. 12, 22539 (2022).
Corbett, A. J., Awad, W., Wang, H. & Chen, Z. Antigen recognition by MR1-reactive T cells; MAIT cells, metabolites, and remaining mysteries. Front. Immunol. 11, 1961 (2020).
Eckle, S. B. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211, 1585–1600 (2014).
Keller, A. N. et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 18, 402–411 (2017). This study reveals that MR1 can present a diverse range of metabolites and drug-like molecules with functional consequences for MAIT cells.
Naidoo, K. et al. MR1‐dependent immune surveillance of the skin contributes to pathogenesis and is a photobiological target of UV light therapy in a mouse model of atopic dermatitis. Allergy 76, 3155–3170 (2021).
Harriff, M. J. et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol. 3, eaao2556 (2018).
Meermeier, E. W. et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat. Commun. 7, 12506 (2016).
Souter, M. N. T. et al. CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells. J. Exp. Med. 219, e20210828 (2022).
Ler, G. J. M. et al. Computer modelling and synthesis of deoxy and monohydroxy analogues of a ribitylaminouracil bacterial metabolite that potently activates human T cells. Chemistry 25, 15594–15608 (2019).
Mak, J. Y. W., Liu, L. & Fairlie, D. P. Chemical modulators of mucosal associated invariant T cells. Acc. Chem. Res. 54, 3462–3475 (2021).
Lange, J. et al. The chemical synthesis, stability, and activity of MAIT cell prodrug agonists that access MR1 in recycling endosomes. ACS Chem. Biol. 15, 437–445 (2020).
Harriff, M. J. et al. Human lung epithelial cells contain Mycobacterium tuberculosis in a late endosomal vacuole and are efficiently recognized by CD8+ T cells. PLoS ONE 9, e97515 (2014).
Chen, Z. et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 10, 58–68 (2017).
Lim, H. J. et al. A specialized tyrosine-based endocytosis signal in MR1 controls antigen presentation to MAIT cells. J. Cell Biol. 221, e202110125 (2022). This study reveals the molecular mechanism of MR1 cell surface display and endocytosis.
Yao, T. et al. MAIT cells accumulate in ovarian cancer-elicited ascites where they retain their capacity to respond to MR1 ligands and cytokine cues. Cancer Immunol. Immunother. 71, 1259–1273 (2021).
Harriff, M. J. et al. Endosomal mr1 trafficking plays a key role in presentation of Mycobacterium tuberculosis ligands to MAIT cells. PLoS Pathog. 12, e1005524 (2016). This study finds several secretory pathway regulators that are required for MR1 antigen presentation.
Le Nours, J. et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366, 1522–1527 (2019).
Wilson, N. S., El-Sukkari, D. & Villadangos, J. A. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood 103, 2187–2195 (2004).
Salio, M. et al. Ligand-dependent downregulation of MR1 cell surface expression. Proc. Natl Acad. Sci. USA 117, 10465–10475 (2020).
Villadangos, J. A., Schnorrer, P. & Wilson, N. S. Control of MHC class II antigen presentation in dendritic cells: a balance between creative and destructive forces. Immunol. Rev. 207, 191–205 (2005).
Huang, S. et al. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205, 1201–1211 (2008).
McWilliam, H. E. G. et al. Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl Acad. Sci. USA 117, 24974–24985 (2020). This study describes a tagged MR1 ligand and uses it to show that MR1 binds extracellular ligands directly in the ER and is stabilized in this compartment by tapasin or TAPBPR.
Abós, B. et al. Human MR1 expression on the cell surface is acid sensitive, proteasome independent and increases after culturing at 26 °C. Biochem. Biophys. Res. Commun. 411, 632–636 (2011).
Yamaguchi, H. & Hashimoto, K. Association of MR1 protein, an MHC class I-related molecule, with β2-microglobulin. Biochem. Biophys. Res. Commun. 290, 722–729 (2002).
Miley, M. J. et al. Biochemical features of the MHC-related protein 1 consistent with an immunological function. J. Immunol. 170, 6090–6098 (2003).
Ljunggren, H. G. et al. Empty MHC class I molecules come out in the cold. Nature 346, 476–480 (1990).
Nelson, C. A., Petzold, S. J. & Unanue, E. R. Peptides determine the lifespan of MHC class II molecules in the antigen-presenting cell. Nature 371, 250–252 (1994).
Sadegh-Nasseri, S., Stern, L. J., Wiley, D. C. & Germain, R. N. MHC class II function preserved by low-affinity peptide interactions preceding stable binding. Nature 370, 647–650 (1994).
Hill, A. et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375, 411–415 (1995).
Kulicke, C. A. et al. The P5-type ATPase ATP13A1 modulates major histocompatibility complex I-related protein 1 (MR1)-mediated antigen presentation. J. Biol. Chem. 298, 101542 (2021). This study uses a genome-wide screen to reveal that MR1 levels and antigen presentation are modulated by the loss of the P5-type ATPase ATP13A1.
McKenna, M. J. et al. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 369, eabc5809 (2020).
Blees, A. et al. Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 (2017).
Wearsch, P. A. & Cresswell, P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin–ERp57 heterodimer. Nat. Immunol. 8, 873–881 (2007).
Jiang, J. et al. Crystal structure of a TAPBPR–MHC-I complex reveals the mechanism of peptide editing in antigen presentation. Science 358, 1064–1068 (2017).
Boyle, L. H. et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc. Natl Acad. Sci. USA 110, 3465–3470 (2013).
Neerincx, A. & Boyle, L. H. Properties of the tapasin homologue TAPBPR. Curr. Opin. Immunol. 46, 97–102 (2017).
Thomas, C. & Tampe, R. Proofreading of peptide–MHC complexes through dynamic multivalent interactions. Front. Immunol. 8, 65 (2017).
Dong, G., Wearsch, P. A., Peaper, D. R., Cresswell, P. & Reinisch, K. M. Insights into MHC class I peptide loading from the structure of the tapasin–ERp57 thiol oxidoreductase heterodimer. Immunity 30, 21–32 (2009).
Thomas, C. & Tampe, R. MHC I assembly and peptide editing—chaperones, clients, and molecular plasticity in immunity. Curr. Opin. Immunol. 70, 48–56 (2021).
Purcell, A. W. et al. Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J. Immunol. 166, 1016–1027 (2001).
McShan, A. C. et al. TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules. Nat. Chem. Biol. 18, 859–868 (2022). This study provides the structural basis for how TAPBPR chaperones MR1.
Bashirova, A. A. et al. HLA tapasin independence: broader peptide repertoire and HIV control. Proc. Natl Acad. Sci. USA 117, 28232–28238 (2020).
Peh, C. A., Laham, N., Burrows, S. R., Zhu, Y. & McCluskey, J. Distinct functions of tapasin revealed by polymorphism in MHC class I peptide loading. J. Immunol. 164, 292–299 (2000).
Ussher, J. E. et al. TLR signalling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur. J. Immunol. 46, 1600–1614 (2016).
Liu, J. & Brutkiewicz, R. R. The Toll-like receptor 9 signalling pathway regulates MR1-mediated bacterial antigen presentation in B cells. Immunol 152, 232–242 (2017).
Villadangos, J. A. et al. MHC class II expression is regulated in dendritic cells independently of invariant chain degradation. Immunity 14, 739–749 (2001).
Vembar, S. S. & Brodsky, J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 (2008).
Purohit, S. K. et al. Varicella zoster virus impairs expression of the non-classical major histocompatibility complex class I-related gene protein (MR1). J. Infect. Dis. 227, 391–401 (2021).
McSharry, B. P. et al. Virus-mediated suppression of the antigen presentation molecule MR1. Cell Rep. 30, 2948–2962.e4 (2020). This study is the first to reveal that viruses can specifically target MR1 for degradation.
Ashley, C. L. et al. Suppression of MR1 by human cytomegalovirus inhibits MAIT cell activation. Front Immunol. 14, 1107497 (2023).
Samer, C. et al. Viral impacts on MR1 antigen presentation to MAIT cells. Crit. Rev. Immunol. 41, 49–67 (2021).
Nikolich-Zugich, J., Slifka, M. K. & Messaoudi, I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4, 123–132 (2004).
Corse, E., Gottschalk, R. A. & Allison, J. P. Strength of TCR–peptide/MHC interactions and in vivo T cell responses. J. Immunol. 186, 5039–5045 (2011).
Adams, B. M., Oster, M. E. & Hebert, D. N. Protein quality control in the endoplasmic reticulum. Protein J. 38, 317–329 (2019).
Caramelo, J. J., Castro, O. A., Alonso, L. G., De Prat-Gay, G. & Parodi, A. J. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc. Natl Acad. Sci. USA 100, 86–91 (2003).
Vassilakos, A., Cohen-Doyle, M. F., Peterson, P. A., Jackson, M. R. & Williams, D. B. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 15, 1495–1506 (1996).
Peaper, D. R. & Cresswell, P. Regulation of MHC class I assembly and peptide binding. Annu. Rev. Cell Dev. Biol. 24, 343–368 (2008).
McWilliam, H. E. G. & Villadangos, J. A. How MR1 presents a pathogen metabolic signature to mucosal-associated invariant T (MAIT) cells. Trends Immunol. 38, 679–689 (2017).
Kumari, S., Mg, S. & Mayor, S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 20, 256–275 (2010).
Doherty, G. J. & McMahon, H. T. Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 (2009).
Ohno, H. et al. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273, 25915–25921 (1998).
Karamooz, E., Harriff, M. J., Narayanan, G. A., Worley, A. & Lewinsohn, D. M. MR1 recycling and blockade of endosomal trafficking reveal distinguishable antigen presentation pathways between Mycobacterium tuberculosis infection and exogenously delivered antigens. Sci. Rep. 9, 4797 (2019).
McWilliam, H. E. & Villadangos, J. A. MR1 antigen presentation to MAIT cells: new ligands, diverse pathways? Curr. Opin. Immunol. 52, 108–113 (2018).
Karamooz, E., Harriff, M. J. & Lewinsohn, D. M. MR1-dependent antigen presentation. Semin. Cell Dev. Biol. 84, 58–64 (2018).
Kulicke, C., Karamooz, E., Lewinsohn, D. & Harriff, M. Covering all the bases: complementary MR1 antigen presentation pathways sample diverse antigens and intracellular compartments. Front. Immunol. 11, 2034 (2020).
McWilliam, H. E. & Villadangos, J. A. MR1: a multi-faceted metabolite sensor for T cell activation. Curr. Opin. Immunol. 64, 124–129 (2020).
Eggensperger, S. & Tampe, R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol. Chem. 396, 1059–1072 (2015).
Sandvig, K. & Van Deurs, B. Transport of protein toxins into cells: pathways used by ricin, cholera toxin and Shiga toxin. FEBS Lett. 529, 49–53 (2002).
Ackerman, A. L., Kyritsis, C., Tampe, R. & Cresswell, P. Access of soluble antigens to the endoplasmic reticulum can explain cross-presentation by dendritic cells. Nat. Immunol. 6, 107–113 (2005).
Zhang, Y. et al. Mucosal-associated invariant T cells restrict reactive oxidative damage and preserve meningeal barrier integrity and cognitive function. Nat. Immunol. 23, 1714–1725 (2022).
Soudais, C. et al. In vitro and in vivo analysis of the Gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J. Immunol. 194, 4641–4649 (2015).
Patel, O. et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142 (2013).
Pentcheva, T., Spiliotis, E. T. & Edidin, M. Cutting edge: tapasin is retained in the endoplasmic reticulum by dynamic clustering and exclusion from endoplasmic reticulum exit sites. J. Immunol. 168, 1538–1541 (2002).
Barnden, M. J., Purcell, A. W., Gorman, J. J. & McCluskey, J. Tapasin-mediated retention and optimization of peptide ligands during the assembly of class I molecules. J. Immunol. 165, 322–330 (2000).
Meyerholz, A. et al. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic 6, 1225–1234 (2005).
Kaksonen, M. & Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018).
Traub, L. M. & Bonifacino, J. S. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5, a016790 (2013).
Dickson, L. J., Liu, S. & Storrie, B. Rab6 is required for rapid, cisternal-specific, intra-Golgi cargo transport. Sci. Rep. 10, 16604 (2020).
Martinez, O. et al. The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol. 127, 1575–1588 (1994).
Martinez, O. et al. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 94, 1828–1833 (1997).
Utskarpen, A., Slagsvold, H. H., Iversen, T.-G., Wälchli, S. & Sandvig, K. Transport of ricin from endosomes to the Golgi apparatus is regulated by Rab6A and Rab6A′. Traffic 7, 663–672 (2006).
Mallard, F. et al. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 (2002).
Spessott, W. A., Sanmillan, M. L., Kulkarni, V. V., McCormick, M. E. & Giraudo, C. G. Syntaxin 4 mediates endosome recycling for lytic granule exocytosis in cytotoxic T-lymphocytes. Traffic 18, 442–452 (2017).
Stow, J. L., Manderson, A. P. & Murray, R. Z. SNAREing immunity: the role of SNAREs in the immune system. Nat. Rev. Immunol. 6, 919–929 (2006).
Ganley, I. G., Espinosa, E. & Pfeffer, S. R. A syntaxin 10–SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 180, 159–172 (2008).
Hirose, H. et al. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23, 1267–1278 (2004).
Hatsuzawa, K. et al. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 275, 13713–13720 (2000).
Williams, D. & Pessin, J. E. Mapping of R-SNARE function at distinct intracellular GLUT4 trafficking steps in adipocytes. J. Cell Biol. 180, 375–387 (2008).
Fritzius, T., Frey, A. D., Schweneker, M., Mayer, D. & Moelling, K. WD-repeat-propeller-FYVE protein, ProF, binds VAMP2 and protein kinase Cζ. FEBS J. 274, 1552–1566 (2007).
Shitara, A. et al. VAMP4 is required to maintain the ribbon structure of the Golgi apparatus. Mol. Cell. Biochem. 380, 11–21 (2013).
Imai, T. et al. Us3 kinase encoded by herpes simplex virus 1 mediates downregulation of cell surface major histocompatibility complex class I and evasion of CD8+ T cells. PLoS ONE 8, e72050 (2013).
Rao, P. et al. Herpes simplex virus 1 glycoprotein B and US3 collaborate to inhibit CD1d antigen presentation and NKT cell function. J. Virol. 85, 8093–8104 (2011).
Seidel, E. et al. Dynamic co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA*008 to escape elimination by NK cells. Cell Rep. 10, 968–982 (2015).
Abendroth, A., Lin, I., Slobedman, B., Ploegh, H. & Arvin, A. M. Varicella-zoster virus retains major histocompatibility complex class I proteins in the Golgi compartment of infected cells. J. Virol. 75, 4878–4888 (2001).
Riegert, P., Wanner, V. & Bahram, S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. 161, 4066–4077 (1998).
Yamaguchi, H., Hirai, M., Kurosawa, Y. & Hashimoto, K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem. Biophys. Res. Commun. 238, 697–702 (1997).
Tsukamoto, K., Deakin, J. E., Graves, J. A. & Hashimoto, K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics 65, 115–124 (2013).
Robinson, J. et al. IPD-IMGT/HLA database. Nucleic Acids Res. 48, D948–D955 (2020).
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Rozemuller, E. et al. MR1 encompasses at least six allele groups with coding region alterations. HLA 98, 509–516 (2021).
Seshadri, C. et al. A polymorphism in human MR1 is associated with mRNA expression and susceptibility to tuberculosis. Genes. Immun. 18, 8–14 (2017).
Robinson, J. et al. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLOS Genet. 13, e1006862 (2017).
Huang, S. et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl Acad. Sci. USA 106, 8290–8295 (2009).
Boudinot, P. et al. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl Acad. Sci. USA 113, E2983–E2992 (2016). This study describes MR1 conservation in mammalian evolution and finds that MR1 and the MAIT cell TCRα chain (TRAV1) are intricately intertwined through evolution.
Acknowledgements
H.E.G.M. discloses support for the research of this work from the National Health and Medical Research Council of Australia (NHMRC) (2003192). J.A.V. discloses support for the research of this work from the Australian Research Council (ARC) (DP170102471), the NHMRC (1113293 and 1058193) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) (R01AI148407).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks Mariolina Salio, Oliver Lantz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
McWilliam, H.E.G., Villadangos, J.A. MR1 antigen presentation to MAIT cells and other MR1-restricted T cells. Nat Rev Immunol 24, 178–192 (2024). https://doi.org/10.1038/s41577-023-00934-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-023-00934-1