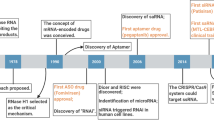
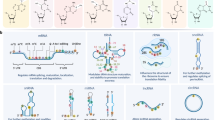
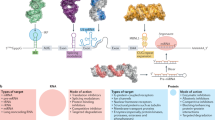
Abstract
The ability of chemical modifications of single nucleotides to alter the electrostatic charge, hydrophobic surface and base pairing of RNA molecules is exploited for the clinical use of stable artificial RNAs such as mRNA vaccines and synthetic small RNA molecules — to increase or decrease the expression of therapeutic proteins. Furthermore, naturally occurring biochemical modifications of nucleotides regulate RNA metabolism and function to modulate crucial cellular processes. Studies showing the mechanisms by which RNA modifications regulate basic cell functions in higher organisms have led to greater understanding of how aberrant RNA modification profiles can cause disease in humans. Together, these basic science discoveries have unravelled the molecular and cellular functions of RNA modifications, have provided new prospects for therapeutic manipulation and have led to a range of innovative clinical approaches.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Amos, H. & Korn, M. 5-Methyl cytosine in the RNA of Escherichia coli. Biochim. Biophys. Acta 29, 444–445 (1958).
Cohn, W. E. Pseudouridine, a carbon–carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J. Biol. Chem. 235, 1488–1498 (1960).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Shi, H. H., Chai, P. W., Jia, R. B. & Fan, X. Q. Novel insight into the regulatory roles of diverse RNA modifications: re-defining the bridge between transcription and translation. Mol. Cancer 19, 78 (2020).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016). This work presents one of the first mechanistic examples showing that inhibition of m5C formation in tRNAs by depleting NSUN2 increases the sensitivity of squamous cell carcinoma cells to chemotherapy in vivo.
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014).
Wilkinson, E., Cui, Y. H. & He, Y. Y. Context-dependent roles of RNA modifications in stress responses and diseases. Int. J. Mol. Sci. 22, 1949 (2021).
Gkatza, N. A. et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 17, e3000297 (2019).
Delaunay, S. & Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 21, 552–559 (2019).
Gattazzo, F., Urciuolo, A. & Bonaldo, P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 1840, 2506–2519 (2014).
Chatterjee, B., Shen, C. J. & Majumder, P. RNA modifications and RNA metabolism in neurological disease pathogenesis. Int. J. Mol. Sci. 22, 11870 (2021).
Thorleifsson, G. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 41, 18–24 (2009).
Frayling, T. M. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 8, 657–662 (2007).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Wu, P. et al. Adaptive mechanisms of tumor therapy resistance driven by tumor microenvironment. Front. Cell Dev. Biol. 9, 641469 (2021).
Holcik, M. & Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 (2005).
Boulias, K. & Greer, E. L. Biological roles of adenine methylation in RNA. Nat. Rev. Genet. 24, 143–160 (2023).
Srinivasan, S., Torres, A. G. & Ribas de Pouplana, L. Inosine in biology and disease. Genes 12, 600 (2021).
Murakami, S. & Jaffrey, S. R. Hidden codes in mRNA: control of gene expression by m6A. Mol. Cell 82, 2236–2251 (2022).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017). This paper reports the first transcriptome-wide mapping of cap m6Am modification, showing that the modification can be removed by FTO and is important for mRNA stability.
Shi, H. L., Wei, J. B. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Wei, J. et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5 (2018). This study shows that cell-specific distribution of the demethylase FTO in the nucleus versus cytoplasm alters its substrate preferences towards demethylating internal m6A modification or cap m6Am modification.
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Zaccara, S. & Jaffrey, S. R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1582–1595.e18 (2020).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Han, D. et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274 (2019).
Zhang, Y. et al. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl Acad. Sci. USA 116, 976–981 (2019).
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311 (2017).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Liu, J. et al. N6-Methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Liu, J. et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591, 322–326 (2021).
Daffis, S. et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456 (2010).
Zust, R. et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 (2011).
Galloway, A. & Cowling, V. H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 270–279 (2019).
Despic, V. & Jaffrey, S. R. mRNA ageing shapes the Cap2 methylome in mammalian mRNA. Nature 614, 358–366 (2023). This study quantifies levels of cap 2′-O-Me at the second transcribed nucleotide (cap2) and finds that cap2 methylation is enriched in long-lived mRNAs and functions to reduce activation of the innate immune response.
Pandey, R. R. et al. The mammalian Cap-specific m6Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 32, 108038 (2020).
Boulias, K. et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75, 631–643.e8 (2019).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Sendinc, E. et al. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e9 (2019).
Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018). This study shows that the N6-adenosine-methyltransferase complex catalytic subunit METTL3 promotes translation of oncogenic mRNAs in the cytoplasm through a mechanism of mRNA looping that involves direct physical and functional interaction between METTL3 and the eukaryotic translation initiation factor eIF3h.
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Coots, R. A. et al. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7 (2017).
Wei, X. et al. METTL3 preferentially enhances non-m6A translation of epigenetic factors and promotes tumourigenesis. Nat. Cell Biol. 24, 1278–1290 (2022).
Delaunay, S. & Frye, M. Localization-dictated function for METTL3. Nat. Cell Biol. 24, 1188–1189 (2022).
Lacerda, R., Menezes, J. & Romao, L. More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell. Mol. Life Sci. 74, 1659–1680 (2017).
Meyer, K. D. et al. 5’ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Doamekpor, S. K., Sharma, S., Kiledjian, M. & Tong, L. Recent insights into noncanonical 5′ capping and decapping of RNA. J. Biol. Chem. 298, 102171 (2022).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Li, X. et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e9 (2017).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Selmi, T. et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 49, 1006–1022 (2021).
Huang, T., Chen, W., Liu, J., Gu, N. & Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26, 380–388 (2019).
Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018).
Wells, S. E., Hillner, P. E., Vale, R. D. & Sachs, A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 (1998).
Schutz, P. et al. Crystal structure of the yeast eIF4A–eIF4G complex: an RNA-helicase controlled by protein–protein interactions. Proc. Natl Acad. Sci. USA 105, 9564–9569 (2008).
Boo, S. H. & Kim, Y. K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 52, 400–408 (2020).
Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Zhou, K. I. et al. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 428, 822–833 (2016).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013).
Porman, A. M. et al. A single N6-methyladenosine site regulates lncRNA HOTAIR function in breast cancer cells. PLoS Biol. 20, e3001885 (2022).
Chen, L. et al. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res. 50, 11619–11634 (2022).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Zhao, Y., Karijolich, J., Glaunsinger, B. & Zhou, Q. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep. 17, 1441–1451 (2016).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Liu, X. & Shan, G. Mitochondria encoded non-coding RNAs in cell physiology. Front. Cell Dev. Biol. 9, 713729 (2021).
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020). This work provides a comprehensive atlas of RNA modifications in human mitochondrial tRNAs and a list of 34 genes responsible for these modifications.
Hopper, A. K. & Nostramo, R. T. tRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front. Genet. 10, 96 (2019).
Kessler, A. C., Silveira d’Almeida, G. & Alfonzo, J. D. The role of intracellular compartmentalization on tRNA processing and modification. RNA Biol. 15, 554–566 (2018).
Hanson, G. & Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018).
Nguyen, H. A., Hoffer, E. D. & Dunham, C. M. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGPro for decoding. J. Biol. Chem. 294, 5281–5291 (2019).
Wang, X. et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 (2018).
Schaefer, M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 (2010).
Tuorto, F. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905 (2012).
Thompson, D. M., Lu, C., Green, P. J. & Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 (2008).
Gebetsberger, J., Wyss, L., Mleczko, A. M., Reuther, J. & Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 14, 1364–1373 (2017).
Kuscu, C. et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 (2018).
Guzzi, N. et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 24, 299–306 (2022).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018). This work describes for the first time how pseudouridylation of tRNA-derived fragments by PUS7 regulates translation in stem cells.
Li, X., Peng, J. & Yi, C. The epitranscriptome of small non-coding RNAs. Noncoding RNA Res. 6, 167–173 (2021).
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-Methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Heo, I. et al. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151, 521–532 (2012).
Pandolfini, L. et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol. Cell 74, 1278–1290.e9 (2019).
Liang, H. et al. 3′-Terminal 2′-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 48, 7027–7040 (2020).
Pendleton, K. E. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14 (2017).
Mendel, M. et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 184, 3125–3142.e25 (2021).
Kim, J. & Lee, G. Metabolic control of m6A RNA modification. Metabolites 11, 80 (2021).
Steurer, B. et al. DNA damage-induced transcription stress triggers the genome-wide degradation of promoter-bound Pol II. Nat. Commun. 13, 3624 (2022).
Buszczak, M., Signer, R. A. & Morrison, S. J. Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159, 242–251 (2014).
Ernens, I. et al. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res. 34, 286–294 (2006).
Coller, J. & Parker, R. General translational repression by activators of mRNA decapping. Cell 122, 875–886 (2005).
Chen, C. Y. & Shyu, A. B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2, 167–183 (2011).
Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019).
Khong, A., Matheny, T., Huynh, T. N., Babl, V. & Parker, R. Limited effects of m6A modification on mRNA partitioning into stress granules. Nat. Commun. 13, 3735 (2022).
Anders, M. et al. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113 (2018).
Ranjan, N. & Rodnina, M. V. tRNA wobble modifications and protein homeostasis. Translation 4, e1143076 (2016).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015).
Llorens-Bobadilla, E. et al. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015).
Signer, R. A., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Blanco, S. et al. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7, e1002403 (2011).
Yue, T. et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 372, eaba4220 (2021).
Szaflarski, W. et al. Early rRNA processing is a stress-dependent regulatory event whose inhibition maintains nucleolar integrity. Nucleic Acids Res. 50, 1033–1051 (2022).
Hannan, K. M. et al. Nuclear stabilization of p53 requires a functional nucleolar surveillance pathway. Cell Rep. 41, 111571 (2022).
Yang, K., Yang, J. & Yi, J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress. 2, 125–140 (2018).
Sloan, K. E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Janin, M. et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 138, 1053–1074 (2019).
Schosserer, M. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 6, 6158 (2015). This work provides the first functional evidence of conserved roles of m5C in rRNA in ageing.
Tower, J. Stress and stem cells. Wiley Interdiscip. Rev. Dev. Biol. 1, 789–802 (2012).
Bednarova, A. et al. Lost in translation: defects in transfer RNA modifications and neurological disorders. Front. Mol. Neurosci. 10, 135 (2017).
Blaze, J. et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavio. Nat. Commun. 12, 4913 (2021). This work directly links NSUN2-mediated m5C in tRNAs to depression-related behaviours in mice.
de Brouwer, A. P. M. et al. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 103, 1045–1052 (2018).
Nostvik, M. et al. Clinical and molecular delineation of PUS3-associated neurodevelopmental disorders. Clin. Genet. 100, 628–633 (2021).
Salvetat, N. et al. A game changer for bipolar disorder diagnosis using RNA editing-based biomarkers. Transl. Psychiatry 12, 182 (2022).
Flores, J. V. et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 8, 112–124 (2017).
Nagayoshi, Y. et al. Loss of Ftsj1 perturbs codon-specific translation efficiency in the brain and is associated with X-linked intellectual disability. Sci. Adv. 7, eabf3072 (2021).
Dewe, J. M., Fuller, B. L., Lentini, J. M., Kellner, S. M. & Fu, D. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell Biol. 37, e00214–e00217 (2017).
Lin, S. et al. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e5 (2018).
Harnett, D. et al. A critical period of translational control during brain development at codon resolution. Nat. Struct. Mol. Biol. 29, 1277–1290 (2022).
Teixeira, F. K. & Lehmann, R. Translational control during developmental transitions. Cold Spring Harb. Perspect. Biol. 11, a032987 (2019).
Statoulla, E., Chalkiadaki, K., Karozis, D. & Gkogkas, C. G. Regulation of mRNA translation in stem cells; links to brain disorders. Cell Signal. 88, 110166 (2021).
Baser, A. et al. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019).
Iwata, R. et al. Mitochondria metabolism sets the species-specific tempo of neuronal development. Science 379, eabn4705 (2023).
Glock, C. et al. The translatome of neuronal cell bodies, dendrites, and axons. Proc. Natl Acad. Sci. USA 118, e2113929118 (2021).
Flamand, M. N. & Meyer, K. D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. 50, 4464–4483 (2022). This work identifies m6A modifications as regulating mRNA localization in hippocampal neurons and shows that the m6A reader proteins YTHDF2 and YTHDF3 mediate this effect.
Richter, J. D. & Zhao, X. Y. The molecular biology of FMRP: new insights into fragile X syndrome. Nat. Rev. Neurosci. 22, 209–222 (2021).
Zhang, F. et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018).
Edupuganti, R. R. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Edens, B. M. et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854.e5 (2019).
Reich, D. P. & Bass, B. L. Mapping the dsRNA world. Cold Spring Harb. Perspect. Biol. 11, a035352 (2019).
Bhogal, B. et al. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat. Neurosci. 14, 1517–1524 (2011).
Shamay-Ramot, A. et al. Fmrp interacts with Adar and regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet. 11, e1005702 (2015).
Verkerk, A. J. et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991).
Tran, S. S. et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25–36 (2019).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016).
Bohnsack, M. T. & Sloan, K. E. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell Mol. Life Sci. 75, 241–260 (2018).
Moraes, C. T., Ricci, E., Bonilla, E., DiMauro, S. & Schon, E. A. The mitochondrial tRNALeu(UUR) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am. J. Hum. Genet. 50, 934–949 (1992).
Suzuki, T., Nagao, A. & Suzuki, T. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip. Rev. RNA 2, 376–386 (2011).
Noer, A. S. et al. A tRNALys mutation in the mtDNA is the causal genetic lesion underlying myoclonic epilepsy and ragged-red fiber (MERRF) syndrome. Am. J. Hum. Genet. 49, 715–722 (1991).
D’Souza, A. R. & Minczuk, M. Mitochondrial transcription and translation: overview. Essays Biochem. 62, 309–320 (2018).
Delaunay, S. et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 607, 593–603 (2022). This work shows that mitochondrial RNA modifications promote the invasive spread of head and neck cancer cells by increasing mRNA translation, leading to increased mitochondrial plasticity and cellular flexibility of metastasis-initiating tumour cells.
Paramasivam, A., Meena, A. K., Venkatapathi, C., Pitceathly, R. D. S. & Thangaraj, K. Novel biallelic NSUN3 variants cause early-onset mitochondrial encephalomyopathy and seizures. J. Mol. Neurosci. 70, 1962–1965 (2020).
Van Haute, L. et al. Deficient methylation and formylation of mt-tRNAMet wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 7, 12039 (2016).
Nakano, S. et al. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol. 12, 546–551 (2016).
Powell, C. A. et al. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am. J. Hum. Genet. 97, 319–328 (2015).
Lee, C., Kramer, G., Graham, D. E. & Appling, D. R. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 282, 27744–27753 (2007).
Ohira, T. & Suzuki, T. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc. Natl Acad. Sci. USA 108, 10502–10507 (2011).
Hoffer, E. D. et al. Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing. eLife 9, e51898 (2020).
Paris, Z. et al. The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 19, 649–658 (2013).
Noma, A., Kirino, Y., Ikeuchi, Y. & Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154 (2006).
Perche-Letuvee, P., Molle, T., Forouhar, F., Mulliez, E. & Atta, M. Wybutosine biosynthesis: structural and mechanistic overview. RNA Biol. 11, 1508–1518 (2014).
Rossello-Tortella, M. et al. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc. Natl Acad. Sci. USA 117, 20785–20793 (2020).
Zhang, L. S. et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat. Cell Biol. 23, 684–691 (2021).
Fu, D., Jordan, J. J. & Samson, L. D. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 27, 1089–1100 (2013).
Kwak, S. H., Park, K. S., Lee, K. U. & Lee, H. K. Mitochondrial metabolism and diabetes. J. Diabetes Investig. 1, 161–169 (2010).
Palmer, C. J. et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 6, 1212–1225 (2017).
Wei, F. Y. et al. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121, 3598–3608 (2011).
Seidel-Rogol, B. L., McCulloch, V. & Shadel, G. S. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem–loop. Nat. Genet. 33, 23–24 (2003).
Liu, X. D. et al. Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function. Nucleic Acids Res. 47, 7648–7665 (2019).
Sharoyko, V. V. et al. Loss of TFB1M results in mitochondrial dysfunction that leads to impaired insulin secretion and diabetes. Hum. Mol. Genet. 23, 5733–5749 (2014).
Koeck, T. et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 13, 80–91 (2011).
Fergus, C., Barnes, D., Alqasem, M. A. & Kelly, V. P. The queuine micronutrient: charting a course from microbe to man. Nutrients 7, 2897–2929 (2015).
Harada, F. & Nishimura, S. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry 11, 301–308 (1972).
Tuorto, F. et al. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37, e99777 (2018).
Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589 (2002). This work provides a major insight into metabolites as constituents of RNA modifications, which has led to clinical applications of high-taurine diets for the treatment of taurine-related human tRNA modopathies (diseases caused by aberrations in tRNA modifications).
Ripps, H. & Shen, W. Review: Taurine: a “very essential” amino acid. Mol. Vis. 18, 2673–2686 (2012).
Singh, P. et al. Taurine deficiency as a driver of aging. Science 380, eabn9257 (2023).
Fakruddin, M. et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 22, 482–496 (2018).
Zeharia, A. et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 85, 401–407 (2009).
Wu, Y. et al. Mtu1-mediated thiouridine formation of mitochondrial tRNAs is required for mitochondrial translation and is involved in reversible infantile liver injury. PLoS Genet. 12, e1006355 (2016).
Kopajtich, R. et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 95, 708–720 (2014).
Ghezzi, D. et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 90, 1079–1087 (2012).
Ohsawa, Y. et al. Taurine supplementation for prevention of stroke-like episodes in MELAS: a multicentre, open-label, 52-week phase III trial. J. Neurol. Neurosurg. Psychiatry 90, 529–536 (2019).
Tomoda, E. et al. Restoration of mitochondrial function through activation of hypomodified tRNAs with pathogenic mutations associated with mitochondrial diseases. Nucleic Acids Res. 2023, gkad139 (2023).
Nutter, C. A. & Kuyumcu-Martinez, M. N. Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wires RNA https://doi.org/10.1002/wrna.1459 (2018).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Scott, L. J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Dina, C. et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726 (2007).
Wang, L. et al. NADP modulates RNA m6A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 16, 1394–1402 (2020).
Fischer, J. et al. Inactivation of the Fto gene protects from obesity. Nature 458, 894–898 (2009).
Gao, X. et al. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE 5, e14005 (2010).
Church, C. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092 (2010).
Ben-Haim, M. S. et al. Dynamic regulation of N6,2′-O-dimethyladenosine (m6Am) in obesity. Nat. Commun. 12, 7185 (2021).
Martinez-Reyes, I. & Chandel, N. S. Cancer metabolism: looking forward. Nat. Rev. Cancer 21, 669–680 (2021).
Begik, O. et al. Integrative analyses of the RNA modification machinery reveal tissue- and cancer-specific signatures. Genome Biol. 21, 97 (2020). This work provides a comprehensive annotation of human RNA-modifying proteins.
Santi, D. V., Garrett, C. E. & Barr, P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 33, 9–10 (1983).
Khan, C., Pathe, N., Fazal, S., Lister, J. & Rossetti, J. M. Azacitidine in the management of patients with myelodysplastic syndromes. Ther. Adv. Hematol. 3, 355–373 (2012).
Lu, L. W., Chiang, G. H., Medina, D. & Randerath, K. Drug effects on nucleic acid modification. I. A specific effect of 5-azacytidine on mammalian transfer RNA methylation in vivo. Biochem. Biophys. Res. Commun. 68, 1094–1101 (1976).
Ladang, A. et al. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 212, 2057–2075 (2015).
Delaunay, S. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 213, 2503–2523 (2016).
Rapino, F. et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609 (2018). This work provides a mechanistic link between defects in tRNA modifications and the mistranslation of a protein target (HIF1), leading to metabolic rewiring and re-sensitization of therapy-resistant melanoma to BRAF inhibitors.
Rosu, A. et al. Loss of tRNA-modifying enzyme Elp3 activates a p53-dependent antitumor checkpoint in hematopoiesis. J. Exp. Med. 218, e20200662 (2021).
Xu, S. W. et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat. Commun. 10, 5492 (2019).
Waszak, S. M. et al. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature 580, 396–401 (2020). This work provides evidence that genetic predisposition to proteome instability caused by germline Elongator complex mutations in Sonic Hedgehog medulloblastoma can be a determining factor in the pathogenesis of paediatric brain cancers.
Vendramin, R. et al. Activation of the integrated stress response confers vulnerability to mitoribosome-targeting antibiotics in melanoma. J. Exp. Med. 218, e20210571 (2021).
Vasan, K., Werner, M. & Chandel, N. S. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 32, 341–352 (2020).
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I. & Klaholz, B. P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017).
Khoshnevis, S., Dreggors-Walker, R. E., Marchand, V., Motorin, Y. & Ghalei, H. Ribosomal RNA 2′-O-methylations regulate translation by impacting ribosome dynamics. Proc. Natl Acad. Sci. USA 119, e2117334119 (2022).
Motorin, Y., Quinternet, M., Rhalloussi, W. & Marchand, V. Constitutive and variable 2′-O-methylation (Nm) in human ribosomal RNA. RNA Biol. 18, 88–97 (2021).
Pauli, C. et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 135, 2059–2070 (2020).
Zhou, F. et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 19, 844–855 (2017).
Zhou, F. et al. A dynamic rRNA ribomethylome drives stemness in acute myeloid leukemia. Cancer Discov. 13, 332–347 (2023). This work provides evidence that a distinct rRNA 2′-O-Me landscape maintains malignant self-renewal in patients with acute myeloid leukaemia.
Butler, M., van der Meer, L. T. & van Leeuwen, F. N. Amino acid depletion therapies: starving cancer cells to death. Trends Endocrinol. Metab. 32, 367–381 (2021).
Kiss-Laszlo, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M. & Kiss, T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077–1088 (1996).
Cavaille, J., Nicoloso, M. & Bachellerie, J. P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383, 732–735 (1996).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021). This work identifies and characterizes the first small-molecule inhibitor (STM2457) targeting the catalytic activity of METTL3, leading to the rapid development of lead compounds in a collaborative setting between biotech and academia.
Paris, J. et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25, 137–148.e6 (2019). This work demonstrates that targeting the m6A reader protein YTHDF2 specifically compromises self-renewing leukaemic stem cells but does not affect normal haematopoietic stem cell function.
Mapperley, C. et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J. Exp. Med. 218, e20200829 (2021).
Esteve-Puig, R. et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood 137, 994–999 (2021).
Chen, X. et al. 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019).
Chen, Z. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47, 2533–2545 (2019).
Legrand, C. et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27, 1589–1596 (2017).
Miao, L., Zhang, Y. & Huang, L. mRNA vaccine for cancer immunotherapy. Mol. Cancer 20, 41 (2021).
Vavilis, T. et al. mRNA in the context of protein replacement therapy. Pharmaceutics 15, 166 (2023).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Freund, I., Eigenbrod, T., Helm, M. & Dalpke, A. H. RNA modifications modulate activation of innate Toll-like receptors. Genes 10, 92 (2019).
Quin, J. et al. ADAR RNA modifications, the epitranscriptome and innate immunity. Trends Biochem. Sci. 46, 758–771 (2021).
Jockel, S. et al. The 2’-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 (2012).
Gehrig, S. et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 (2012).
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008). This work shows the benefits of incorporating RNA modifications into medicinal mRNA by in vitro transcription.
Andries, O. et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Rel. 217, 337–344 (2015).
Anderson, B. R. et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010).
Svitkin, Y. V. et al. N1-Methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036 (2017).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Kormann, M. S. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011).
Svitkin, Y. V., Gingras, A. C. & Sonenberg, N. Membrane-dependent relief of translation elongation arrest on pseudouridine- and N1-methyl-pseudouridine-modified mRNAs. Nucleic Acids Res. 50, 7202–7215 (2022). This work provides important novel mechanistic insights into the question of how RNA modifications modulate translation elongation, a feature that has greatly benefitted the development of mRNA vaccines.
Mei, Y. & Wang, X. RNA modification in mRNA cancer vaccines. Clin. Exp. Med. https://doi.org/10.1007/s10238-023-01020-5 (2023).
Mauro, V. P. Codon optimization in the production of recombinant biotherapeutics: potential risks and considerations. BioDrugs 32, 69–81 (2018).
Kim, Y. K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 54, 455–465 (2022).
Crooke, S. T., Vickers, T. A. & Liang, X. H. Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 48, 5235–5253 (2020).
Hua, Y. et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010).
Lefebvre, S. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 (1995).
Hua, Y., Vickers, T. A., Okunola, H. L., Bennett, C. F. & Krainer, A. R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82, 834–848 (2008).
Amanat, M., Nemeth, C. L., Fine, A. S., Leung, D. G. & Fatemi, A. Antisense oligonucleotide therapy for the nervous system: from bench to bedside with emphasis on pediatric neurology. Pharmaceutics 14, 2389 (2022).
Zhang, X., Goel, V. & Robbie, G. J. Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J. Clin. Pharmacol. 60, 573–585 (2020).
Scott, L. J. Givosiran: first approval. Drugs 80, 335–339 (2020).
Hu, B. et al. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 5, 101 (2020).
Temple, G. F., Dozy, A. M., Roy, K. L. & Kan, Y. W. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature 296, 537–540 (1982).
Lueck, J. D. et al. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 10, 822 (2019).
Ko, W., Porter, J. J., Sipple, M. T., Edwards, K. M. & Lueck, J. D. Efficient suppression of endogenous CFTR nonsense mutations using anticodon-engineered transfer RNAs. Mol. Ther. Nucleic Acids 28, 685–701 (2022).
Albers, S. et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 618, 842–848 (2023).
Karijolich, J. & Yu, Y. T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011).
Adachi, H. et al. Targeted pseudouridylation: an approach for suppressing nonsense mutations in disease genes. Mol. Cell 83, 637–651.e9 (2023).
Song, J. et al. CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons. Mol. Cell 83, 139–155.e9 (2023).
Gort, L., Chabas, A. & Coll, M. J. Analysis of five mutations in 20 mucopolysaccharidois type 1 patients: high prevalence of the W402X mutation. Mutations in brief no. 121. Hum. Mutat. 11, 332–333 (1998).
Presnyak, V. et al. Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 (2015).
Schaffrath, R. & Leidel, S. A. Wobble uridine modifications — a reason to live, a reason to die?! RNA Biol. 14, 1209–1222 (2017).
Hou, Y. M., Masuda, I. & Gamper, H. Codon-specific translation by m1G37 methylation of tRNA. Front. Genet. 9, 713 (2018).
Lamichhane, T. N. et al. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol. Cell Biol. 33, 2918–2929 (2013).
Ivanov, P., Emara, M. M., Villen, J., Gygi, S. P. & Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011).
Torres, A. G., Reina, O., Stephan-Otto Attolini, C. & Ribas de Pouplana, L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl Acad. Sci. USA 116, 8451–8456 (2019).
Choe, B. K. & Taylor, M. W. Kinetics of synthesis and characterization of transfer-RNA precursors in mammalian cells. Biochim. Biophys. Acta 272, 275–287 (1972).
Werner, M. et al. 2’-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 39, 4756–4768 (2011).
Belanger, F., Stepinski, J., Darzynkiewicz, E. & Pelletier, J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 285, 33037–33044 (2010).
Uzonyi, A. et al. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol. Cell 83, 237–251.e7 (2022).
He, P. C. et al. Exon architecture controls mRNA m6A suppression and gene expression. Science 379, 677–682 (2023).
Yang, X., Triboulet, R., Liu, Q., Sendinc, E. & Gregory, R. I. Exon junction complex shapes the m6A epitranscriptome. Nat. Commun. 13, 7904 (2022).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 5, e18434 (2016).
Su, S. et al. Cryo-EM structures of human m6A writer complexes. Cell Res. 32, 982–994 (2022).
Bawankar, P. et al. Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat. Commun. 12, 3778 (2021).
Haag, S. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 35, 2104–2119 (2016).
Thuring, K., Schmid, K., Keller, P. & Helm, M. LC-MS analysis of methylated RNA. Methods Mol. Biol. 1562, 3–18 (2017).
Wiener, D. & Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 22, 119–131 (2021).
Motorin, Y. & Helm, M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes 10, 35 (2019).
Cui, J., Liu, Q., Sendinc, E., Shi, Y. & Gregory, R. I. Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res. 49, e27 (2021).
Zhou, H. et al. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods 16, 1281–1288 (2019).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9, e110799 (2014).
Carlile, T. M., Rojas-Duran, M. F. & Gilbert, W. V. Transcriptome-wide identification of pseudouridine modifications using Pseudo-seq. Curr. Protoc. Mol. Biol. 112, 4.25.1–4.25.24 (2015).
Dai, Q. et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat. Biotechnol. 41, 344–354 (2023).
Sugimoto, Y. et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein–RNA interactions. Genome Biol. 13, R67 (2012).
Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280 (2019).
Begik, O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 39, 1278–1291 (2021). This study uses nanopore RNA direct sequencing to detect the RNA modifications m6A, Ψ and 2′-O-Me in cellular RNAs.
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Jain, M., Olsen, H. E., Akeson, M. & Abu-Shumays, R. Adaptation of human ribosomal RNA for nanopore sequencing of canonical and modified nucleotides. Methods Mol. Biol. 2298, 53–74 (2021).
Leger, A. et al. RNA modifications detection by comparative nanopore direct RNA sequencing. Nat. Commun. 12, 7198 (2021).
Relier, S. et al. Multivariate analysis of RNA chemistry marks uncovers epitranscriptomics-based biomarker signature for adult diffuse glioma diagnostics. Anal. Chem. 94, 11967–11972 (2022).
Zhou, B. et al. RNA modification writer expression profiles predict clinical outcomes and guide neoadjuvant immunotherapy in non-small cell lung cancer. EBioMedicine 84, 104268 (2022).
Zhang, P. et al. Scoring system based on RNA modification writer-related genes to predict overall survival and therapeutic response in bladder cancer. Front. Immunol. 12, 724541 (2021).
Heidenreich, O., Pieken, W. & Eckstein, F. Chemically modified RNA: approaches and applications. FASEB J. 7, 90–96 (1993).
Soffer, D., Stoekenbroek, R. & Plakogiannis, R. Small interfering ribonucleic acid for cholesterol lowering—inclisiran: inclisiran for cholesterol lowering. J. Clin. Lipidol. 16, 574–582 (2022).
Manoharan, M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 8, 570–579 (2004).
Bumcrot, D., Manoharan, M., Koteliansky, V. & Sah, D. W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2, 711–719 (2006).
Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid. Ther. 24, 374–387 (2014).
Kellner, S. et al. Oxidation of phosphorothioate DNA modifications leads to lethal genomic instability. Nat. Chem. Biol. 13, 888–894 (2017).
Wu, Y. et al. RNA phosphorothioate modification in prokaryotes and eukaryotes. ACS Chem. Biol. 15, 1301–1305 (2020).
Kaiser, S. et al. Strategies to avoid artifacts in mass spectrometry-based epitranscriptome analyses. Angew. Chem. Int. Ed. Engl. 60, 23885–23893 (2021).
Acknowledgements
M.F. received funding from the Helmholtz Association (W2/W3-106), Cancer Research UK (CR-UK; C10701/A15181) and Worldwide Cancer Research (21-0223). S.D. was supported by an EMBO (European Molecular Biology Organization) long-term fellowship (LTFS48) and by the Leon Fredericq Foundation. M.H. and M.F. are part of TRR319 RMaP (439669440).
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, contributed substantially to discussion of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
Research in the group of M.F. is partly funded by Merck. M.H. is a consultant for Moderna, Inc. S.D. declares no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks the anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ClinicalTrials.gov: https://www.clinicaltrials.gov
Glossary
- Anticodon
-
A sequence of three nucleotides on a tRNA molecule that recognizes and binds to a complementary trinucleotide codon sequence on mRNA during protein synthesis.
- Antisense oligonucleotides
-
(ASOs). Short synthetic strands of nucleic acids that bind to complementary RNA sequences, offering targeted gene regulation and therapeutic potential for various genetic and disease-related applications.
- Base stacking
-
Non-covalent interaction between adjacent aromatic nitrogenous bases in RNA that contributes to the stability of the secondary and tertiary structures of the RNA.
- Charged tRNAs
-
tRNA molecules that carry a specific amino acid and are ready to participate in protein synthesis during translation.
- Codon optimality
-
Refers to the preferential use of certain synonymous codons over others in a given organism or gene owing to differences in their usage frequency or interactions with tRNA molecules and ribosomes.
- Elongator complex
-
A multi-protein complex composed of two subcomplexes, ELP1–ELP2–ELP3 and ELP4–ELP5–ELP6, that modifies tRNAs in their wobble position to regulate protein synthesis and ensure proteome stability.
- Fragile X syndrome
-
A genetic disorder characterized by developmental delays, learning disabilities and social and behavioural problems, caused by a mutation in FMR1, which is needed for brain development.
- Mitochondrial respiratory chain complex deficiency
-
A type of mitochondrial disease caused by defects in the enzymes involved in oxidative phosphorylation (OXPHOS), resulting in impaired energy production.
- Oxidative phosphorylation
-
(OXPHOS). A metabolic pathway taking place inside mitochondria, in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in the form of ATP.
- Queuosine
-
A hypermodified guanosine present in certain tRNAs in bacteria and eukaryotes containing the nucleobase queuine, which has a role in maintaining the proper reading frame during mRNA translation.
- Unfolded protein response
-
A cellular stress response mechanism that is activated by the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum.
- Wobble position
-
The third nucleotide position of the anticodon trinucleotide sequence, which pairs with more than one complementary nucleotide in the mRNA codon.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Delaunay, S., Helm, M. & Frye, M. RNA modifications in physiology and disease: towards clinical applications. Nat Rev Genet 25, 104–122 (2024). https://doi.org/10.1038/s41576-023-00645-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-023-00645-2
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
Epitranscriptomic modifications in mesenchymal stem cell differentiation: advances, mechanistic insights, and beyond
Cell Death & Differentiation (2024)
-
Expedient production of site specifically nucleobase-labelled or hypermodified RNA with engineered thermophilic DNA polymerases
Nature Communications (2024)