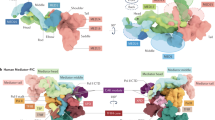
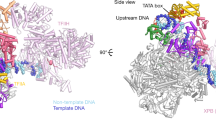
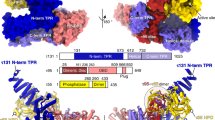
Abstract
The RNA polymerase II (Pol II) pre-initiation complex (PIC) is a critical node in eukaryotic transcription regulation, and its formation is the major rate-limiting step in transcriptional activation. Diverse cellular signals borne by transcriptional activators converge on this large, multiprotein assembly and are transduced via intermediary factors termed coactivators. Cryogenic electron microscopy, multi-omics and single-molecule approaches have recently offered unprecedented insights into both the structure and cellular functions of the PIC and two key PIC-associated coactivators, Mediator and TFIID. Here, we review advances in our understanding of how Mediator and TFIID interact with activators and affect PIC formation and function. We also discuss how their functions are influenced by their chromatin environment and selected cofactors. We consider how, through its multifarious interactions and functionalities, a Mediator-containing and TFIID-containing PIC can yield an integrated signal processing system with the flexibility to determine the unique temporal and spatial expression pattern of a given gene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Roeder, R. G. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp. Quant. Biol. 63, 201–218 (1998).
Thomas, M. C. & Chiang, C. M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006).
Schier, A. C. & Taatjes, D. J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488 (2020).
Roeder, R. G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 (1996).
Buratowski, S. et al. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334, 37–42 (1988).
Horikoshi, M. et al. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature 341, 299–303 (1989).
Hoey, T. et al. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell 61, 1179–1186 (1990).
Kao, C. C. et al. Cloning of a transcriptionally active human TATA binding factor. Science 248, 1646–1650 (1990).
Kostrewa, D. et al. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
He, Y. et al. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013). This seminal study uses cryo-EM to visualize intermediates in the assembly pathway of a TBP-nucleated PIC that has previously been biochemically described.
He, Y. et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016).
Plaschka, C. et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016).
Murakami, K. et al. Structure of an RNA polymerase II preinitiation complex. Proc. Natl Acad. Sci. USA 112, 13543–13548 (2015).
Nogales, E. & Greber, B. J. High-resolution cryo-EM structures of TFIIH and their functional implications. Curr. Opin. Struct. Biol. 59, 188–194 (2019).
Aibara, S., Schilbach, S. & Cramer, P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 594, 124–128 (2021).
Schilbach, S. et al. Structure of RNA polymerase II pre-initiation complex at 2.9 Å defines initial DNA opening. Cell 184, 4064–4072 e28 (2021). Together with Aibara et al. (2021), this work provides high-resolution structural views of PICs that have been trapped in intermediate states and sheds new light on the mechanism of promoter melting through the TFIIH translocase.
Pal, M., Ponticelli, A. S. & Luse, D. S. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 (2005).
Pugh, B. F. & Tjian, R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell 61, 1187–1197 (1990).
Hoffman, A. et al. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 346, 387–390 (1990).
Goodrich, J. A. & Tjian, R. TBP–TAF complexes: selectivity factors for eukaryotic transcription. Curr. Opin. Cell Biol. 6, 403–409 (1994).
Malik, S. & Roeder, R. G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25, 277–283 (2000).
Myers, L. C. & Kornberg, R. D. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69, 729–749 (2000).
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137 (2000).
Guermah, M., Malik, S. & Roeder, R. G. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell Biol. 18, 3234–3244 (1998).
Baek, H. J. et al. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell Biol. 22, 2842–2852 (2002).
Black, J. C. et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23, 809–818 (2006).
Grunberg, S. et al. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 35, 2435–2446 (2016).
Sun, F. et al. The Pol II preinitiation complex (PIC) influences Mediator binding but not promoter–enhancer looping. Genes Dev. 35, 1175–1189 (2021).
Richter, W. F. et al. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell. Biol. 23, 732–749 (2022).
Malik, S. & Roeder, R. G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010).
Allen, B. L. & Taatjes, D. J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015).
Tsai, K. L. et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 (2014). This paper provides the first detailed insights into subunit and modular organization of the Mediator core complex by a combination of cryo-EM, biochemical and genetic approaches.
Toth-Petroczy, A. et al. Malleable machines in transcription regulation: the Mediator complex. PLoS Comput. Biol. 4, e1000243 (2008).
Wang, X. et al. Redefining the modular organization of the core Mediator complex. Cell Res. 24, 796–808 (2014).
Cevher, M. A. et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 (2014). In this study, a core human Mediator complex that retains many of its effector functions is recombinantly generated, allowing assessment of roles of individual subunits, including a pivotal architectural and functional role for MED14.
El Khattabi, L. et al. A pliable Mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158.e20 (2019).
Plaschka, C., Nozawa, K. & Cramer, P. Mediator architecture and RNA polymerase II interaction. J. Mol. Biol. 428, 2569–2574 (2016).
Robinson, P. J. et al. Molecular architecture of the yeast Mediator complex. eLife 4, e08719 (2015).
Robinson, P. J. et al. Structure of a complete Mediator–RNA polymerase II pre-initiation complex. Cell 166, 1411–1422.e16 (2016). This study is one of the first to describe a Mediator-containing PIC combining cryo-EM and cross-linking–mass spectroscopy-based integrative modelling.
Nozawa, K., Schneider, T. R. & Cramer, P. Core mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545, 248–251 (2017).
Tsai, K. L. et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017).
Zhao, H. et al. Structure of mammalian Mediator complex reveals tail module architecture and interaction with a conserved core. Nat. Commun. 12, 1355 (2021).
Chen, X. et al. Structures of the human mediator and mediator-bound preinitiation complex. Science 372, eabg0635 (2021). This landmark study reveals, using cryo-EM, the most complete views to date of the mammalian PIC assembled with TFIID, Mediator, Pol II and all the GTFs; it allows direct assessment of how the PIC is impacted by its two associated coactivators.
Rengachari, S. et al. Structure of the human mediator–RNA polymerase II pre-initiation complex. Nature 594, 129–133 (2021).
Abdella, R. et al. Structure of the human Mediator-bound transcription preinitiation complex. Science 372, 52–56 (2021). Together with Rengacahari et al. (2021), this work provides structural insights into how Mediator impacts the PIC, albeit in the context of TBP.
Zhang, H. et al. Mediator structure and conformation change. Mol. Cell 81, 1781–1788.e4 (2021).
Louder, R. K. et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016).
Patel, A. B. et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018). Together with Louder et al. (2016), this work first reveals how TFIID engages with the various elements that constitute a core promoter and the dramatic conformational changes it undergoes during the process.
Chen, X. et al. Structural insights into preinitiation complex assembly on core promoters. Science 372, eaba8490 (2021). This important cryo-EM study of TFIID promoter complexes with diverse core promoters further reveals interesting promoter sequence-dependent dynamics and variations in how TFIID interacts with the promoter.
Cianfrocco, M. A. et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152, 120–131 (2013).
Papai, G. et al. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577, 711–716 (2020).
Wang, H. et al. Structure of the transcription coactivator SAGA. Nature 577, 717–720 (2020).
Timmers, H. T. M. SAGA and TFIID: friends of TBP drifting apart. Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194604 (2021).
Deato, M. D. & Tjian, R. An unexpected role of TAFs and TRFs in skeletal muscle differentiation: switching core promoter complexes. Cold Spring Harb. Symp. Quant. Biol. 73, 217–225 (2008).
Hoey, T. et al. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72, 247–260 (1993).
Moqtaderi, Z. et al. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383, 188–191 (1996).
Walker, S. S. et al. Transcription activation in cells lacking TAFIIS. Nature 383, 185–188 (1996).
Timmers, H. T. M. & Tora, L. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol. Cell 72, 10–17 (2018).
Warfield, L. et al. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol. Cell 68, 118–129.e5 (2017).
Wright, K. J. & Tjian, R. Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo. Proc. Natl Acad. Sci. USA 106, 55–60 (2009).
Chen, W. Y. et al. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 27, 1596–1609 (2013). This study provides evidence for TFIID coactivator functions via direct activator–TAF interactions in the context of the native TFIID complex.
Zhang, J. et al. E protein silencing by the leukemogenic AML1–ETO fusion protein. Science 305, 1286–1289 (2004).
Liu, W. L. et al. Structures of three distinct activator–TFIID complexes. Genes Dev. 23, 1510–1521 (2009).
Malik, S. et al. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell Biol. 25, 2117–2129 (2005).
Knoll, E. R. et al. Role of the pre-initiation complex in Mediator recruitment and dynamics. eLife 7, e39633 (2018).
Warfield, L. et al. Yeast Mediator facilitates transcription initiation at most promoters via a tail-independent mechanism. Mol. Cell 82, 4033–4048.e7 (2022).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018).
Yang, F. et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006).
Stevens, J. L. et al. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296, 755–758 (2002).
Ge, K. et al. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417, 563–567 (2002).
Stumpf, M. et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl Acad. Sci. USA 103, 18504–18509 (2006).
Chu, C. S. et al. Unique immune cell coactivators specify locus control region function and cell stage. Mol. Cell 80, 845–861.e10 (2020).
Lee, Y. L. et al. Mediator subunit MED1 is required for E2A-PBX1-mediated oncogenic transcription and leukemic cell growth. Proc. Natl Acad. Sci. USA 118, e1922864118 (2021).
Chen, W., Yang, Q. & Roeder, R. G. Dynamic interactions and cooperative functions of PGC-1α and MED1 in TRα-mediated activation of the brown-fat-specific UCP-1 gene. Mol. Cell 35, 755–768 (2009).
Iida, S. et al. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. 29, 308–321 (2015).
Harms, M. J. et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307 (2015).
Ito, M. et al. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5, 683–693 (2000).
Belorusova, A. Y. et al. Molecular determinants of MED1 interaction with the DNA bound VDR–RXR heterodimer. Nucleic Acids Res. 48, 11199–11213 (2020).
Lee, H. K. et al. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 26, 3545–3557 (2007).
Han, E. H., Rha, G. B. & Chi, Y. I. MED25 is a mediator component of HNF4α-driven transcription leading to insulin secretion in pancreatic β-cells. PLoS ONE 7, e44007 (2012).
Milbradt, A. G. et al. Structure of the VP16 transactivator target in the Mediator. Nat. Struct. Mol. Biol. 18, 410–415 (2011).
Vojnic, E. et al. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18, 404–409 (2011).
Currie, S. L. et al. ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J. Mol. Biol. 429, 2975–2995 (2017).
Tuttle, L. M. et al. Gcn4–Mediator specificity is mediated by a large and dynamic fuzzy protein–protein complex. Cell Rep. 22, 3251–3264 (2018).
Taatjes, D. J. et al. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 (2002).
Hu, Z. et al. A matter of access. nucleosome disassembly from gene promoters is the central goal transcriptional activators. Transcription 5, e29355 (2014).
Cairns, B. R. The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198 (2009).
Jacobson, R. H. et al. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Lauberth, S. M. et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036 (2013).
Matangkasombut, O. et al. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14, 951–962 (2000).
Andrews, F. H. et al. The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription 7, 14–20 (2016).
Nagai, S. et al. Chromatin potentiates transcription. Proc. Natl Acad. Sci. USA 114, 1536–1541 (2017).
Zhu, X. et al. Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 39, 8342–8354 (2011).
Chen, X. et al. Structures of +1 nucleosome-bound PIC–Mediator complex. Science 378, 62–68 (2022).
Nock, A. et al. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48, 837–848 (2012).
Zaret, K. S. Pioneer transcription factors initiating gene network changes. Annu. Rev. Genet. 54, 367–385 (2020).
Workman, J. L. & Roeder, R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51, 613–622 (1987).
Joo, Y. J. et al. Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes Dev. 31, 2162–2174 (2017).
Teves, S. S. et al. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 (2018).
Murakami, S., Nagari, A. & Kraus, W. L. Dynamic assembly and activation of estrogen receptor α enhancers through coregulator switching. Genes Dev. 31, 1535–1548 (2017).
Blobel, G. A. et al. Testing the super-enhancer concept. Nat. Rev. Genet. 22, 749–755 (2021).
Sabari, B. R., Dall’Agnese, A. & Young, R. A. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Shrinivas, K. et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561.e7 (2019).
Zamudio, A. V. et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell 76, 753–766.e6 (2019).
Boehning, M. et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 (2018).
Guo, Y. E. et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019).
Trojanowski, J. et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e10 (2022).
Li, J. et al. Single-molecule nanoscopy elucidates RNA polymerase II transcription at single genes in live cells. Cell 178, 491–506.e28. (2019).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Petrenko, N. et al. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell 64, 443–454 (2016).
Jeronimo, C. et al. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell 64, 455–466 (2016).
Malik, S. & Roeder, R. G. Mediator: a drawbridge across the enhancer–promoter divide. Mol. Cell 64, 433–434 (2016).
Jaeger, M. G. et al. Selective Mediator dependence of cell-type-specifying transcription. Nat. Genet. 52, 719–727 (2020). This comprehensive genetic analysis of the Mediator in human cells uses rapid degradation approaches to monitor how individual subunits impact various transcriptional parameters.
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017).
Karr, J. P. et al. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 36, 7–16 (2022).
McNally, J. G. et al. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000).
Murakami, K. et al. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J. Biol. Chem. 288, 6325–6332 (2013).
Baek, I. et al. Single-molecule studies reveal branched pathways for activator-dependent assembly of RNA polymerase II pre-initiation complexes. Mol. Cell 81, 3576–3588.e6 (2021). This study uses single-molecule microscopy to reveal a novel branched PIC assembly pathway in which TFIIE and TFIIF may preassemble with enhancer-bound activators prior to entry into the PIC.
Gorbea Colon, J. J. et al. Structural basis of a transcription pre-initiation complex on a divergent promoter. Mol. Cell 83, 574–588.e11 (2023).
Rhee, H. S. & Pugh, B. F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012).
Haberle, V. & Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637 (2018).
Plaschka, C. et al. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518, 376–380 (2015).
Schilbach, S. et al. Yeast PIC–Mediator structure with RNA polymerase II C-terminal domain. Proc. Natl Acad. Sci. USA 120, e2220542120 (2023).
Fishburn, J. et al. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl Acad. Sci. USA 112, 3961–3966 (2015).
Kim, T. K., Ebright, R. H. & Reinberg, D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000).
Zhang, Z. et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 30, 2106–2118 (2016).
Baek, H. J., Kang, Y. K. & Roeder, R. G. Human mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 281, 15172–15181 (2006).
Jishage, M. et al. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat. Struct. Mol. Biol. 25, 859–867 (2018).
Malik, S., Molina, H. & Xue, Z. PIC activation through functional interplay between mediator and TFIIH. J. Mol. Biol. 429, 48–63 (2017).
Sogaard, T. M. & Svejstrup, J. Q. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 282, 14113–14120 (2007).
Wong, K. H., Jin, Y. & Struhl, K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell 54, 601–612 (2014).
Fant, C. B. et al. TFIID enables RNA polymerase II promoter-proximal pausing. Mol. Cell 78, 785–793.e8 (2020).
Core, L. & Adelman, K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982 (2019).
Yadav, D. et al. Multivalent role of human TFIID in recruiting elongation components at the promoter-proximal region for transcriptional control. Cell Rep. 26, 1303–1317.e7 (2019).
Takahashi, H. et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011).
Takahashi, H. et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 6, 5941 (2015).
Galbraith, M. D. et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339 (2013).
Sato, S. et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 (2004).
Ranish, J. A., Yudkovsky, N. & Hahn, S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999).
Sikorski, T. W. & Buratowski, S. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 21, 344–351 (2009).
Tantale, K. et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 7, 12248 (2016).
Ehrensberger, A. H., Kelly, G. P. & Svejstrup, J. Q. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell 154, 713–715 (2013). This work is a highly useful mathematical resource that allows assessment of the relative contributions of transcription initiation per se versus subsequent Pol II pausing to overall activity of a given gene.
Eychenne, T. et al. Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev. 30, 2119–2132 (2016).
Li, Y. C. et al. Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Sci. Adv. 7, eabd4484 (2021).
Di Lello, P. et al. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22, 731–740 (2006).
Rochette-Egly, C. et al. Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell 90, 97–107 (1997).
Lin, Y. S. et al. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353, 569–571 (1991).
Nakadai, T. et al. Two target gene activation pathways for orphan ERR nuclear receptors. Cell Res. 33, 165–183 (2023).
Tyree, C. M. et al. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 7, 1254–1265 (1993).
Parvin, J. D. & Sharp, P. A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73, 533–540 (1993).
Goodrich, J. A. & Tjian, R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 77, 145–156 (1994).
Holstege, F. C. et al. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 14, 810–819 (1995).
Dienemann, C. et al. Promoter distortion and opening in the RNA polymerase II cleft. Mol. Cell 73, 97–106.e4 (2019).
Kouzine, F. et al. Global regulation of promoter melting in naive lymphocytes. Cell 153, 988–999 (2013).
Tora, L. & Timmers, H. T. The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem. Sci. 35, 309–314 (2010).
Wu, S. Y. & Chiang, C. M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282, 13141–13145 (2007).
van Werven, F. J. et al. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369 (2008).
Hsu, J. Y. et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353–2358 (2008).
Bhagwat, A. S. et al. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 15, 519–530 (2016).
Sdelci, S. et al. Mapping the chemical chromatin reactivation landscape identifies BRD4–TAF1 cross-talk. Nat. Chem. Biol. 12, 504–510 (2016).
Donczew, R. & Hahn, S. BET family members Bdf1/2 modulate global transcription initiation and elongation in Saccharomyces cerevisiae. eLife 10, e69619 (2021).
Acknowledgements
The authors apologize to colleagues whose work could not be cited directly owing to space constraints. The authors thank past and current members of their laboratory for their many contributions to the authors’ understanding of the pre-initiation complex (PIC) and its coactivators. The authors are thankful to M. Gnädig, K. Ito and T. Onikubo for critical reading of the manuscript. The authors’ work was supported by National Institutes of Health (NIH) grants CA234575, CA273709 and AI148387.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Eva Nogales and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
This article is dedicated to the memory of Michael R. Green.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- +1 Nucleosome
-
A precisely positioned nucleosome located just downstream from the core promoter. Together with an upstream localized −1 nucleosome, it delimits the boundaries of a nucleosome-depleted region. It is not to be confused with the transcription start site (TSS), which is sometimes also referred to as the +1 site.
- Basal transcription
-
Baseline levels of transcription that might occur in the absence of activation signals.
- Bromodomain
-
A conserved protein domain that can recognize acetylated lysine residues. It is found in many chromatin readers, as well as BET (bromodomain and extra-terminal) family members such as BRD4.
- Core promoter
-
A regulatory region within a transcriptional locus that consists of numerous elements that collectively or singly specify the transcription start site (TSS) by recruiting and orienting the pre-initiation complex (PIC). Elements include the TATA box, initiator (Inr), motif ten element (MTE) and downstream promoter element (DPE), among others.
- Enhancers
-
Regulatory elements that carry binding sites for multiple transcriptional activators. Typically, enhancers are found at distal locations relative to the transcription start site (TSS), although some are located more proximally. Variants include super-enhancers, which are characterized by a large number of activator binding sites. In yeast, the upstream activator binding site (UAS) serves the same purpose.
- Initiation
-
The formation of the first phosphodiester bond by pre-initiation complex (PIC)-associated RNA polymerase II (Pol II) following ATP hydrolysis-dependent promoter melting.
- Intrinsically disordered regions
-
(IDRs). Regions in a protein that lack any defined three-dimensional structure, at least in the absence of an interacting protein. They have a high propensity to form higher-order structures through weak multivalent interactions.
- Pausing
-
Transient stalling of transcribing RNA polymerase II (Pol II) soon after promoter clearance that is regulated by a combination of pause-inducing and pause-release factors.
- Phase separation
-
A tendency of two immiscible liquids to separate into distinct phases. In the transcription field, the term largely refers to the tendency of some activators and coactivators that contain intrinsically disordered regions (IDRs) to form condensates when forced into situations where they can cluster, for example on super-enhancers.
- Pol II stalk
-
A prominent feature of RNA polymerase II (Pol II) composed mainly of RPB4 and RPB7 subunits. It can regulate other Pol II elements (for example, the clamp) and acts as a hub for the interaction of multiple pre-initiation complex (PIC) components.
- Promoter escape
-
A series of post-initiation events that occur as a prelude to RNA polymerase II (Pol II) entry into the elongation mode and result in relinquishing of stabilizing interactions that anchor the pre-initiation complex (PIC). This process is closely related to promoter clearance.
- RPB1 CTD
-
The large unstructured carboxy-terminal domain (CTD) of the largest RNA polymerase II (Pol II) subunit. In human, it consists of 52 heptapeptide repeats with the consensus sequence YSPTSPS. The repeat units are subject to phosphorylation at multiple residues. In the context of the pre-initiation complex (PIC), Ser5 is the predominant target of the cyclin-dependent kinase (CDK)-activating kinase (CAK) module of TFIIH.
- YEATS domain
-
One of several domains that can recognize acylated lysine residues. So called because of its original identification in the chromatin reading modules of Yaf9, ENL, AF9, TAF1 and Sas5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malik, S., Roeder, R.G. Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators. Nat Rev Genet 24, 767–782 (2023). https://doi.org/10.1038/s41576-023-00630-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-023-00630-9