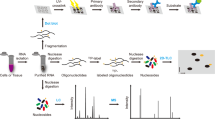
Abstract
Chemical modifications to nucleic acids occur across the kingdoms of life and carry important regulatory information. Reliable high-resolution mapping of these modifications is the foundation of functional and mechanistic studies, and recent methodological advances based on next-generation sequencing and long-read sequencing platforms are critical to achieving this aim. However, mapping technologies may have limitations that sometimes lead to inconsistent results. Some of these limitations are technical in nature and specific to certain types of technology. Here, however, we focus on common (yet not always widely recognized) pitfalls that are shared among frequently used mapping technologies and discuss strategies to help technology developers and users mitigate their effects. Although the emphasis is primarily on DNA modifications, RNA modifications are also discussed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019).
Michalak, E. M., Burr, M. L., Bannister, A. J. & Dawson, M. A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 20, 573–589 (2019).
Jiang, X. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6, 74 (2021).
Sánchez-Romero, M. A. & Casadesús, J. The bacterial epigenome. Nat. Rev. Microbiol. 18, 7–20 (2020). This review summarizes epigenetic regulation by bacterial DNA methylation and its contribution to phenotypic heterogeneity in bacterial populations.
Luo, C., Hajkova, P. & Ecker, J. R. Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340 (2018).
Wu, X. & Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018).
Beaulaurier, J., Schadt, E. E. & Fang, G. Deciphering bacterial epigenomes using modern sequencing technologies. Nat. Rev. Genet. 20, 157–172 (2019). This review discusses the potential of currently available methods, especially LRS technologies, for mapping and characterizing bacterial methylomes.
Blow, M. J. et al. The epigenomic landscape of prokaryotes. PLoS Genet. 12, 1–28 (2016).
Boulias, K. & Greer, E. L. Means, mechanisms and consequences of adenine methylation in DNA. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00456-x (2022).
Fang, G. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239 (2012). This paper applies SMRT sequencing to map 6mA in a bacterium at genome-wide scale and highlights the importance of FDR evaluation.
Beaulaurier, J. et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat. Biotechnol. 36, 61–69 (2018).
Kumar, S. & Mohapatra, T. Deciphering epitranscriptome: modification of mRNA bases provides a new perspective for post-transcriptional regulation of gene expression. Front. Cell Dev. Biol. 9, 1–22 (2021).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017). This review discusses the principles, advantages and drawbacks of new high-throughput methods for characterizing RNA modifications.
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Grozhik, A. V. & Jaffrey, S. R. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 14, 215–225 (2018).
Saletore, Y. et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012).
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 6, 863–865 (2010).
Xue, C. et al. Role of main RNA modifications in cancer: N6-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct. Target. Ther. 7, 142 (2022).
Tretyakova, N., Villalta, P. W. & Kotapati, S. Mass spectrometry of structurally modified DNA. Chem. Rev. 113, 2395–2436 (2013).
Boulias, K. & Greer, E. L. in DNA Modifications: Methods and Protocols (eds Ruzov, A. & Gering, M.) 79–90 (Springer US, 2021).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Flusberg, B. A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010). This paper provides an early description of direct mapping of 5mC, 5hmC and 6mA using SMRT sequencing.
Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Laszlo, A. H. et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl Acad. Sci. USA 110, 18904–18909 (2013).
Amarasinghe, S. L. et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 1–16 (2020).
Krueger, F., Kreck, B., Franke, A. & Andrews, S. R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 9, 145–151 (2012).
Darst, R. P., Pardo, C. E., Ai, L., Brown, K. D. & Kladde, M. P. Bisulfite sequencing of DNA. Curr. Protoc. Mol. Biol. https://doi.org/10.1002/0471142727.mb0709s91 (2010).
Shi, D. Q., Ali, I., Tang, J. & Yang, W. C. New insights into 5hmC DNA modification: generation, distribution and function. Front. Genet. 8, 1–11 (2017).
Amente, S. et al. Genome-wide mapping of genomic DNA damage: methods and implications. Cell. Mol. Life Sci. 78, 6745–6762 (2021).
Rybin, M. J. et al. Emerging technologies for genome-wide profiling of DNA breakage. Front. Genet. 11, 610386 (2021).
Zhao, L. Y., Song, J., Liu, Y., Song, C. X. & Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 11, 792–808 (2020). This review summarizes high-throughput NGS-based methods for mapping five forms of DNA modifications and eight forms of RNA modifications, and also summarizes biological discoveries made using these methods.
O’Brown, Z. K. et al. Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genomics 20, 1–15 (2019). This article reports sources of artefacts in measurements of 6mA and 4mC abundance in eukaryotic gDNA, including both quantification methods and mapping methods.
Kong, Y. et al. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 375, 515–522 (2022). This paper describes a machine learning method that can quantitatively deconvolve 6mA events into eukaryotic species of interest and other sources, and warns about bacterial contamination in the study of 6mA in eukaryotic samples.
Patil, V. et al. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 47, 10072–10085 (2019).
Bellizzi, D. et al. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 20, 537–547 (2013).
Dou, X. et al. The strand-biased mitochondrial DNA methylome and its regulation by DNMT3A. Genome Res. 29, 1622–1634 (2019).
Sharma, N., Pasala, M. S. & Prakash, A. Mitochondrial DNA: epigenetics and environment. Environ. Mol. Mutagen. 60, 668–682 (2019).
Owa, C., Poulin, M., Yan, L. & Shioda, T. Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: sources and avoidance of false-positive detection. PLoS ONE 13, 1–19 (2018).
Bicci, I., Calabrese, C., Golder, Z. J., Gomez-Duran, A. & Chinnery, P. F. Single-molecule mitochondrial DNA sequencing shows no evidence of CpG methylation in human cells and tissues. Nucleic Acids Res. 49, 12757–12768 (2021). This paper describes the bias and technical concerns related to BS-seq in mapping 5mC in mtDNA and reports more reliable 5mC levels estimated by machine learning modelling of nanopore data.
Mechta, M., Ingerslev, L. R., Fabre, O., Picard, M. & Barrès, R. Evidence suggesting absence of mitochondrial DNA methylation. Front. Genet. 8, 1–9 (2017).
Matsuda, S. et al. Accurate estimation of 5-methylcytosine in mammalian mitochondrial DNA. Sci. Rep. 8, 1–13 (2018).
Hong, E. E., Okitsu, C. Y., Smith, A. D. & Hsieh, C.-L. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell. Biol. 33, 2683–2690 (2013).
Cao, K., Feng, Z., Gao, F., Zang, W. & Liu, J. Mitoepigenetics: an intriguing regulatory layer in aging and metabolic-related diseases. Free. Radic. Biol. Med. 177, 337–346 (2021).
Stoccoro, A. & Coppedè, F. Mitochondrial DNA methylation and human diseases. Int. J. Mol. Sci. 22, 1–27 (2021).
Lopes, A. F. C. Mitochondrial metabolism and DNA methylation: a review of the interaction between two genomes. Clin. Epigenetics 12, 1–13 (2020).
Zhang, G. et al. N6-Methyladenine DNA modification in Drosophila. Cell 161, 893–906 (2015).
Wu, T. P. et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016).
Greer, E. L. et al. DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878 (2015).
Zhou, C. et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants 4, 554–563 (2018).
Hao, Z. et al. N6-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell https://doi.org/10.1016/j.molcel.2020.02.018 (2020).
Wang, Y., Chen, X., Sheng, Y., Liu, Y. & Gao, S. N6-Adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45, 11594–11606 (2017).
Fu, Y. et al. N6-Methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Xie, Q. et al. N6-Methyladenine DNA modification in glioblastoma. Cell 175, 1228–1243.e20 (2018).
Xiao, C. L. et al. N6-Methyladenine DNA modification in the human genome. Mol. Cell 71, 306–318.e7 (2018).
Lentini, A. et al. A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 15, 499–504 (2018). This paper demonstrates systematic off-target binding of antibodies to unmodified short tandem repeats in DIP-seq studies and highlights the importance of using matched IgG negative controls.
Douvlataniotis, K., Bensberg, M., Lentini, A., Gylemo, B. & Nestor, C. E. No evidence for DNA N6-methyladenine in mammals. Sci. Adv. 6, 1–10 (2020). This article reports that DIP-seq libraries can be contaminated with mammalian mRNA, and that high abundance of m6A in mRNA can contribute to false positive 6mA peaks in DIP-seq.
Musheev, M. U., Baumgärtner, A., Krebs, L. & Niehrs, C. The origin of genomic N6-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 16, 630–634 (2020).
Foox, J. et al. The SEQC2 epigenomics quality control (EpiQC) study. Genome Biol. 22, 1–30 (2021).
Skvortsova, K. et al. Comprehensive evaluation of genome-wide 5-hydroxymethylcytosine profiling approaches in human DNA. Epigenetics Chromatin 10, 1–20 (2017).
Branco, M. R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2012).
Olova, N. et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 19, 1–19 (2018).
Ramsahoye, B. H. et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA 97, 5237–5242 (2000).
Patil, V., Ward, R. L. & Hesson, L. B. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 9, 823–828 (2014).
Jang, H. S., Shin, W. J., Lee, J. E. & Do, J. T. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes 8, 2–20 (2017).
Zhang, Y. et al. Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc. Natl Acad. Sci. USA 115, E1069–E1074 (2018).
Morrison, J. et al. Evaluation of whole-genome DNA methylation sequencing library preparation protocols. Epigenetics Chromatin 14, 1–15 (2021).
Robertson, K. D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005).
Suzuki, M. M. & Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 (2008).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37, e12 (2009).
Legrand, C. et al. Statistically robust methylation calling for whole transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27, 1589–1596 (2017).
Huang, T., Chen, W., Liu, J., Gu, N. & Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26, 380–388 (2019).
Schaefer, M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 560, 297–329 (2015).
Trixl, L., Rieder, D., Amort, T. & Lusser, A. in Epitranscriptomics (eds Wajapeyee, N. & Gupta, R.) 1–21 (Springer, 2019).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Wiener, D. & Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 22, 119–131 (2021). This Perspective discusses the technical and analytical challenges that led to inconsistent conclusions regarding the abundance and distribution of six forms of RNA modifications beyond m6A.
Luo, G. Z. et al. Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 7, 2–7 (2016).
Cohen-Karni, D. et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl Acad. Sci. USA 108, 11040–11045 (2011).
Sun, Z. et al. A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell 57, 750–761 (2015).
Roberts, R. J., Vincze, T., Posfai, J. & Macelis, D. REBASE — a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015).
Zhang, Z. et al. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5, 1–12 (2019).
Garcia-Campos, M. A. et al. Deciphering the “m6A Code” via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019).
Zhang, Z. et al. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods 18, 1213–1222 (2021).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Jin, H., Huo, C., Zhou, T. & Xie, S. m1A RNA modification in gene expression regulation. Genes 13, 910 (2022).
Wei, J. et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5 (2018).
Cui, X. et al. 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol. Plant. 10, 1387–1399 (2017).
Ma, J. et al. M5C-Atlas: a comprehensive database for decoding and annotating the 5-methylcytosine (m5C) epitranscriptome. Nucleic Acids Res. 50, D196–D203 (2022).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Gokhale, N. S. et al. N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016).
Delatte, B. et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25, 679–684 (2011).
Li, W., Li, X., Ma, X., Xiao, W. & Zhang, J. Mapping the m1A, m5C, m6A and m7G methylation atlas in zebrafish brain under hypoxic conditions by MeRIP-seq. BMC Genomics 23, 1–19 (2022).
Song, C. X., Yi, C. & He, C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 30, 1107–1116 (2012).
Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020).
Thalalla Gamage, S., Sas-Chen, A., Schwartz, S. & Meier, J. L. Quantitative nucleotide resolution profiling of RNA cytidine acetylation by ac4C-seq. Nat. Protoc. 16, 2286–2307 (2021).
Grozhik, A. V. et al. Antibody cross-reactivity accounts for widespread appearance of m1A in 5′UTRs. Nat. Commun. 10, 1–13 (2019).
Zhu, S. et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 28, 1067–1078 (2018). This article demonstrates that sequencing technologies tend to make false positive 6mA calls in eukaryotes, and highlights the importance of FDR evaluation for low-abundance modifications.
Liu, X. et al. N6-Methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 31, 94–97 (2021).
Liu, B., Liu, X., Lai, W. & Wang, H. Metabolically generated stable isotope-labeled deoxynucleoside code for tracing DNA N6-methyladenine in human cells. Anal. Chem. 89, 6202–6209 (2017).
O’Brown, Z. K. & Greer, E. L. in DNA methyltransferases — Role and Function (eds Jeltsch, A. & Jurkowska, R. Z.) 213–246 (Springer International, 2016).
Schiffers, S. et al. Quantitative LC-MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. — Int. Ed. 56, 11268–11271 (2017).
Tan, S. C. & Yiap, B. C. DNA, RNA, and protein extraction: the past and the present. J. Biomed. Biotechnol. 2009, 574398 (2009).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9, e1003602 (2013).
Thu, K. L. et al. Methylated DNA immunoprecipitation. J. Vis. Exp. https://doi.org/10.3791/935 (2009).
Amort, T. & Lusser, A. in RNA Methylation (ed. Lusser, A.) 107–121 (Springer, 2017).
Meyer, C. A. & Liu, X. S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat. Rev. Genet. 15, 709–721 (2014).
Sims, D., Sudbery, I., Ilott, N. E., Heger, A. & Ponting, C. P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15, 121–132 (2014).
McIntyre, A. B. R. et al. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 10, 1–15 (2020).
Beaulaurier, J. et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6, 7438 (2015).
Schadt, E. E. et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 (2013).
Vilfan, I. D. et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J. Nanobiotechnol. 11, 1 (2013).
Abdi, E., Latifi-Navid, S. & Latifi-Navid, H. Long noncoding RNA polymorphisms and colorectal cancer risk: progression and future perspectives. Environ. Mol. Mutagen. 63, 98–112 (2022).
Pratanwanich, P. N. et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 39, 1394–1402 (2021).
Jenjaroenpun, P. et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 49, 1–13 (2021).
Price, A. M. et al. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 11, 6016 (2020).
Parker, M. T. et al. Nanopore direct RNA sequencing maps the complexity of arabidopsis mRNA processing and m6A modification. Elife 9, 1–35 (2020).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 1–9 (2019).
Kimura, S., Dedon, P. C. & Waldor, M. K. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 16, 964–972 (2020).
Ameur, A., Kloosterman, W. P. & Hestand, M. S. Single-molecule sequencing: towards clinical applications. Trends Biotechnol. 37, 72–85 (2019).
Fang, L. et al. DeepRepeat: direct quantification of short tandem repeats on signal data from nanopore sequencing. Genome Biol. 23, 1–27 (2022).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Huang, Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, 1–9 (2010).
Liu, Y. et al. DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation. Genome Biol. 22, 295 (2021). This paper benchmarks computational methods for detecting 5mC in mammalian genomes using nanopore sequencing, and demonstrates that 5hmC levels may contribute to the discrepancy between BS-seq and nanopore sequencing.
Clark, T. A., Spittle, K. E., Turner, S. W. & Korlach, J. Direct detection and sequencing of damaged DNA bases. Genome Integr. 2, 10 (2011).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Sun, H. et al. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat. Commun. 12, 1–12 (2021).
Guiblet, W. M. et al. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 28, 1767–1778 (2018).
Kwok, C. K., Tang, Y., Assmann, S. M. & Bevilacqua, P. C. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 40, 221–232 (2015).
Mortimer, S. A., Kidwell, M. A. & Doudna, J. A. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15, 469–479 (2014).
Wan, Y., Kertesz, M., Spitale, R. C., Segal, E. & Chang, H. Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 12, 641–655 (2011).
Talkish, J., May, G., Lin, Y., Woolford, J. L. & McManus, C. J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. Rna 20, 713–720 (2014).
Weeks, K. M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20, 295–304 (2010).
Helm, M., Lyko, F. & Motorin, Y. Limited antibody specificity compromises epitranscriptomic analyses. Nat. Commun. 10, 9–11 (2019).
Zaringhalam, M. & Papavasiliou, F. N. Pseudouridylation meets next-generation sequencing. Methods 107, 63–72 (2016).
Palazzo, A. F. & Lee, E. S. Non-coding RNA: what is functional and what is junk? Front. Genet. 5, 1–11 (2015).
Stark, R., Grzelak, M. & Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656 (2019).
Ozsolak, F. & Milos, P. M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98 (2011).
Hussain, S., Aleksic, J., Blanco, S., Dietmann, S. & Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14, 1–10 (2013).
Oliveira, P. H. et al. Epigenomic characterization of Clostridioides difficile finds a conserved DNA methyltransferase that mediates sporulation and pathogenesis. Nat. Microbiol. 5, 166–180 (2020).
Hong, T. et al. Selective detection of N6-methyladenine in DNA via metal ion-mediated replication and rolling circle amplification. Chem. Sci. 8, 200–205 (2016).
Engel, J. D. & Von Hippel, P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J. Biol. Chem. 253, 927–934 (1978).
Clark, T. A. et al. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 11, 4 (2013).
Chaveza, L. et al. Simultaneous sequencing of oxidized methylcytosines produced by TET/JBP dioxygenases in Coprinopsis cinerea. Proc. Natl Acad. Sci. USA 111, E5149–E5158 (2014).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). This paper describes the use of synthetically methylated DNA to train a hidden Markov model for distinguishing 5mC from unmethylated cytosine in mammalian genomes using nanopore data.
Rand, A. C. et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017).
Tourancheau, A., Mead, E. A., Zhang, X. S. & Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 18, 491–498 (2021). This paper reports that the nanopore sequencing signal displays complex heterogeneity across methylation events of the same type, and develops a method for de novo detection of three forms of bacterial DNA methylation across diverse sequence contexts.
Ni, P. et al. Genome-wide detection of cytosine methylations in plant from nanopore data using deep learning. Nat. Commun. 12, 1–11 (2021).
Liu, Y. et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019).
Liu, Y. et al. Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS. Genome Biol. 21, 1–9 (2020).
Song, C. X. et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat. Methods 9, 75–77 (2012).
Xiong, X., Li, X., Wang, K. & Yi, C. Perspectives on topology of the human m1A methylome at single nucleotide resolution. Rna 24, 1437–1442 (2018).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Gallego Romero, I., Pai, A. A., Tung, J. & Gilad, Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 12, 1–13 (2014).
Schuierer, S. et al. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics 18, 1–13 (2017).
Siwek, W., Czapinska, H., Bochtler, M., Bujnicki, J. M. & Skowronek, K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 40, 7563–7572 (2012).
Mierzejewska, K. et al. Structural basis of the methylation specificity of R.DpnI. Nucleic Acids Res. 42, 8745–8754 (2014).
Kazrani, A. A., Kowalska, M., Czapinska, H. & Bochtler, M. Crystal structure of the 5hmC specific endonuclease PvuRts1I. Nucleic Acids Res. 42, 5929–5936 (2014).
Ni, P. et al. DeepSignal: detecting DNA methylation state from nanopore sequencing reads using deep-learning. Bioinformatics 35, 4586–4595 (2019).
Liu, Q. et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 (2019).
McIntyre, A. B. R. et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 10, 1–11 (2019).
Yuen, Z. W. S. et al. Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing. Nat. Commun. 12, 1–12 (2021).
Zhang, Y. Z. et al. On the application of BERT models for nanopore methylation detection. Proc. 2021 IEEE Int. Conf. Bioinforma. Biomed. BIBM 2021 https://doi.org/10.1109/BIBM52615.2021.9669841 (2021).
Liu, Q., Georgieva, D. C., Egli, D. & Wang, K. NanoMod: a computational tool to detect DNA modifications using nanopore long-read sequencing data. BMC Genomics 20, 78 (2019).
Tse, O. Y. O. et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl Acad. Sci. USA 118, 1–11 (2021).
Ni, P. et al. DNA 5-methylcytosine detection and methylation phasing using PacBio circular consensus sequencing. Preprint at bioRxiv https://doi.org/10.1101/2022.02.26.482074 (2022).
Garalde, D. R. et al. Highly parallel direct RN A sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018). This paper provides an early description of direct RNA sequencing for detecting RNA modifications using the nanopore platform.
Xu, L. & Seki, M. Recent advances in the detection of base modifications using the nanopore sequencer. J. Hum. Genet. 65, 25–33 (2020).
Begik, O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 39, 1278–1291 (2021).
Lorenz, D. A., Sathe, S., Einstein, J. M. & Yeo, G. W. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28 (2020).
Gao, Y. et al. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using nanopore direct RNA sequencing. Genome Biol. 22, 1–17 (2021).
Qin, H. et al. DENA: training an authentic neural network model using nanopore sequencing data of Arabidopsis transcripts for detection and quantification of N6-methyladenosine on RNA. Genome Biol. 23, 1–23 (2022).
Leger, A. et al. RNA modifications detection by comparative nanopore direct RNA sequencing. Nat. Commun. 12, 1–17 (2021).
Whalen, S., Schreiber, J., Noble, W. S. & Pollard, K. S. Navigating the pitfalls of applying machine learning in genomics. Nat. Rev. Genet. 23, 169–181 (2022).
Schutsky, E. K. et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol. 36, 1083–1090 (2018).
Zhang, W., Qian, Y. & Jia, G. The detection and functions of RNA modification m6A based on m6A writers and erasers. J. Biol. Chem. 297, 100973 (2021).
Tavakoli, S. et al. Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct and long-read sequencing. Preprint at bioRxiv https://doi.org/10.1101/2021.11.03.467190 (2022).
Makhamreh, A. et al. Messenger-RNA modification standards and machine learning models facilitate absolute site-specific pseudouridine quantification. Preprint at bioRxiv https://doi.org/10.1101/2022.05.06.490948 (2022).
Plongthongkum, N., Diep, D. H. & Zhang, K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet. 15, 647–661 (2014). This review discusses the principles as well as experimental and analytical challenges in mapping 5mC and its oxidation derivatives with BS-seq and other NGS-based methods.
Lio, C. W. J. & Rao, A. TET enzymes and 5hMC in adaptive and innate immune systems. Front. Immunol. 10, 1–13 (2019).
Lai, W., Lyu, C. & Wang, H. Vertical ultrafiltration-facilitated DNA digestion for rapid and sensitive UHPLC-MS/MS detection of DNA modifications. Anal. Chem. 90, 6859–6866 (2018).
Liu, X., Lai, W., Zhang, N. & Wang, H. Predominance of N6-methyladenine-specific DNA fragments enriched by multiple immunoprecipitation. Anal. Chem. 90, 5546–5551 (2018).
Debo, B. M., Mallory, B. & Stergachis, A. B. Evaluation of N6-adenine DNA-immunoprecipitation-based genomic profiling in eukaryotes. Preprint at bioRxiv https://doi.org/10.1101/2022.03.02.482749 (2022).
Liang, Z. et al. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell 45, 406–416.e3 (2018).
Luo, G. Z. et al. N6-Methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 19, 1–12 (2018).
Mondo, S. J. et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. https://doi.org/10.1038/ng.3859 (2017).
Vaisvila, R. et al. Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 31, 1280–1289 (2021).
Sun, Z. et al. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. 31, 291–300 (2021).
Feng, S., Zhong, Z., Wang, M. & Jacobsen, S. E. Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing. Epigenetics Chromatin 13, 1–17 (2020).
Wang, Y. et al. A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res. 47, 11771–11789 (2019).
Beh, L. Y. et al. Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell 177, 1781–1796.e25 (2019).
Rodriguez, F., Yushenova, I. A., DiCorpo, D. & Arkhipova, I. R. Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA. Nat. Commun. 13, 1–17 (2022).
Dietzsch, J., Feineis, D. & Höbartner, C. Chemoselective labeling and site-specific mapping of 5-formylcytosine as a cellular nucleic acid modification. FEBS Lett. 592, 2032–2047 (2018).
Song, C. X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Zhu, C. et al. Single-cell 5-formylcytosine landscapes of mammalian early embryos and ESCs at single-base resolution. Cell Stem Cell 20, 720–731.e5 (2017).
Shen, L. et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 (2013).
Neri, F., Incarnato, D., Krepelova, A., Parlato, C. & Oliviero, S. Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis. Nat. Protoc. 11, 1191–1205 (2016).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Booth, M. J., Marsico, G., Bachman, M., Beraldi, D. & Balasubramanian, S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6, 435–440 (2014).
Booth, M. J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Füllgrabe, J. et al. Accurate simultaneous sequencing of genetic and epigenetic bases in DNA. Preprint at bioRxiv https://doi.org/10.1101/2022.07.08.499285 (2022).
Shen, L., Song, C. X., He, C. & Zhang, Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614 (2014).
Wescoe, Z. L., Schreiber, J. & Akeson, M. Nanopores discriminate among five C5-cytosine variants in DNA. J. Am. Chem. Soc. 136, 16582–16587 (2014).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Krutyhołowa, R., Zakrzewski, K. & Glatt, S. Charging the code — tRNA modification complexes. Curr. Opin. Struct. Biol. 55, 138–146 (2019).
Sloan, K. E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Decatur, W. A. & Fournier, M. J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
Sergiev, P. V., Aleksashin, N. A., Chugunova, A. A., Polikanov, Y. S. & Dontsova, O. A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 14, 226–235 (2018).
Anreiter, I., Mir, Q., Simpson, J. T., Janga, S. C. & Soller, M. New twists in detecting mRNA modification dynamics. Trends Biotechnol. 39, 72–89 (2021).
Michaela, F., Bryan, T. H., Mikaela, B. & Chuan, H. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Stephenson, W. et al. Direct detection of RNA modifications and structure using single-molecule nanopore sequencing. Cell Genomics 2, 100097 (2022).
Yu, F. et al. CFEA: a cell-free epigenome atlas in human diseases. Nucleic Acids Res. 48, D40–D44 (2020).
Li, W. et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 27, 1243–1257 (2017).
Tian, X. et al. Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res. 28, 597–600 (2018).
Siejka-Zielińska, P. et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 7, eabh0534 (2021).
Johnson, P., Zhou, Q., Dao, D. Y. & Lo, Y. M. D. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 19, 670–681 (2022).
Poetsch, A. R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 18, 207–219 (2020).
Chatterjee, N. & Walker, G. C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263 (2017).
Huang, R. & Zhou, P. K. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 6, 254 (2021).
Abdulhay, N. J. et al. Massively multiplex single-molecule oligonucleosome footprinting. eLife 9, 1–23 (2020).
Shipony, Z. et al. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat. Methods 17, 319–327 (2020).
Wang, Y. et al. Single-molecule long-read sequencing reveals the chromatin basis of gene expression. Genome Res. 29, 1329–1342 (2019).
Stergachis, A. B., Debo, B. M., Haugen, E., Churchman, L. S. & Stamatoyannopoulos, J. A. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science 368, 1449–1454 (2020).
Altemose, N. et al. DiMeLo-seq: a long-read, single-molecule method for mapping protein–DNA interactions genome wide. Nat. Methods 19, 711–723 (2022).
Ghut, J. et al. Determination of isoform-specific RNA structure with nanopore long reads. Nat. Biotechnol. 39, 336–346 (2020).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Pomraning, K. R., Smith, K. M. & Freitag, M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47, 142–150 (2009).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–404 (2011).
Xu, Y. et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 (2011).
Liu, J. et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 7, 1–7 (2016).
Koziol, M. J. et al. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 23, 24–30 (2016).
Ando, Y. & Hayashizaki, Y. Restriction landmark genomic scanning. Nat. Protoc. 1, 2774–2783 (2007).
Koike, K., Matsuyama, T. & Ebisuzaki, T. Epigenetics: application of virtual image restriction landmark genomic scanning (Vi-RLGS). FEBS J. 275, 1608–1616 (2008).
Suzuki, M. & Greally, J. M. DNA methylation profiling using HpaII tiny fragment enrichment by ligation-mediated PCR (HELP). Methods 52, 218–222 (2010).
Oda, M. & Greally, J. M. in DNA Methylation (ed. Tost, J.) 77–87 (Humana, 2009).
Sun, Z. et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 3, 567–576 (2013).
Jia, Z. et al. A 5-mC dot blot assay quantifying the DNA methylation level of chondrocyte dedifferentiation in vitro. J. Vis. Exp. 2017, 5–9 (2017).
Jin, S. G., Wu, X., Li, A. X. & Pfeifer, G. P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 39, 5015–5024 (2011).
Inoue, A., Shen, L., Dai, Q., He, C. & Zhang, Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21, 1670–1676 (2011).
Wang, Y. et al. Naphthalimide derivatives as multifunctional molecules for detecting 5-formylpyrimidine by both PAGE analysis and dot-blot assays. Chem. Commun. 54, 1497–1500 (2018).
Wheldon, L. M. et al. Transient accumulation of 5-carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells. Cell Rep. 7, 1353–1361 (2014).
Shen, L., Liang, Z. & Yu, H. Dot blot analysis of N6-methyladenosine RNA modification levels. Bio-Protoc. 7, 4–8 (2017).
Nagarajan, A., Janostiak, R. & Wajapeyee, N. in Epitranscriptomics (eds Wajapeyee, N. & Gupta, R.) 263–271 (Springer, 2019).
Mishima, E. et al. Immuno-northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS ONE 10, 1–17 (2015).
Shen, C., Wang, K., Deng, X. & Chen, J. DNA N6-methyldeoxyadenosine in mammals and human disease. Trends Genet. 38, 454–467 (2022).
Yu, M. et al. Base-resolution detection of N4-methylcytosine in genomic DNA using 4mC-Tet-assisted-bisulfitesequencing. Nucleic Acids Res. 43, 1–10 (2015).
Faulk, C. in Toxicoepigenetics (eds McCullough, S. D. & Dolinoy, D. C.) 153–171 (Academic Press, 2018).
Acknowledgements
The work was funded by R01 HG011095 (G.F.) and R35 GM139655 (G.F.) from the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content and reviewed and edited the manuscript before submission. Y.K. and G.F. wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Endogenous DNA and RNA modifications
-
DNA and RNA modifications generated during metabolic processes in living organisms, usually catalysed by specific enzymes within the cells.
- Exogenous DNA and RNA modifications
-
DNA and RNA modifications generated by exogenous factors that originated outside the cells, such as externally modified nucleotides that are randomly incorporated during DNA replication, or events that are directly catalysed by exogenous DNA methyltransferases in vitro or in vivo.
- False discovery rates
-
(FDRs). These mathematical concepts describe the expected ratio of the number of false positive classifications to the total number of positive classifications. In the context of mapping DNA or RNA modifications, the FDR refers to the probability of false positive calls among the detected modification events by a mapping method.
- False negative
-
This mathematical concept describes when a test result incorrectly indicates the absence of a condition. In the context of mapping DNA or RNA modifications, it refers to the case whereby an authentic modification event of interest is classified as either unmodified or as a different type of modification.
- False negative rate
-
This mathematical concept describes the probability of making a false negative call for a particular test. In the context of mapping DNA or RNA modifications, it refers to the proportion of false negative calls among all the authentic modifications of interest by a mapping method.
- False positive
-
This mathematical concept describes when a test result incorrectly indicates the presence of a condition. In the context of mapping DNA or RNA modifications, it refers to the case whereby a base is called as modified even though it is not, or a different type of modification is called as the specific modification type of interest.
- False positive rate
-
(FPR). This mathematical concept describes the probability of making false positive calls with a particular test. In the context of mapping DNA or RNA modifications, it refers to the proportion of false positive modification calls among unmodified bases (or modified bases of other types) by a mapping method.
- Heteroplasmic
-
The presence of more than one type of organellar genome (mitochondrial DNA (mtDNA) or plastid DNA) within a cell or individual.
- Inter-pulse duration ratio
-
(IPD ratio). The IPD ratio is the deviation of an observed IPD (the time length between emission pulses associated with base incorporation events) from the expected IPD associated with modification-free DNA with the same flanking sequence context. The IPD ratio reflects the presence of a chemical modification of a nucleotide or its neighbouring nucleotides.
- Methylation motifs
-
Short sequence patterns (usually 2 ~ 10 bp) that are enriched for a certain type of DNA or RNA methylation event in an organism, and which are often determined by the recognition preference of DNA or RNA methyltransferases. For example, >95% of adenines at GATC sites are methylated (N6-methyladenine (6mA)) in γ-proteobacteria, and >80% of cytosines at CpG sites are methylated (5-methylcytosine (5mC)) in the human genome.
- Modification quality value
-
The –log10 transformed p value. In single-molecule, real-time (SMRT) sequencing, the modification quality value describes the significance of the observed inter-pulse duration (IPD) deviation from the expected level (free of modification).
- Multiple hypothesis testing
-
In statistics, the multiple testing problem occurs when a set of statistical inferences based on the observed values are considered simultaneously. The more inferences are made, the more likely erroneous inferences become.
- Restriction-modification systems
-
Rudimentary immune systems found in bacteria and other prokaryotic organisms, which provide defence against foreign DNA. They include a restriction enzyme, which cuts specific unmethylated DNA sequences, and the methyltransferase, which protects the same DNA sequences.
- Sensitivity
-
Also known as the recall or true positive rate, this mathematical concept describes the probability that a positive test is truly positive. In the context of mapping DNA or RNA modifications, it refers to the probability that true modification events are successfully detected as such by a mapping method.
- Specificity
-
Also known as the true negative rate, this mathematical concept describes the probability that a negative test is truly negative, and is expressed as specificity = 1 – false positive rate (FPR). In the context of mapping DNA or RNA modifications, it refers to the probability that a modified event detected by a mapping method truly belongs to the modification type of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, Y., Mead, E.A. & Fang, G. Navigating the pitfalls of mapping DNA and RNA modifications. Nat Rev Genet 24, 363–381 (2023). https://doi.org/10.1038/s41576-022-00559-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00559-5