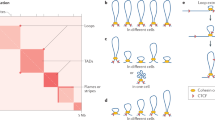
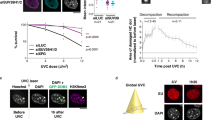
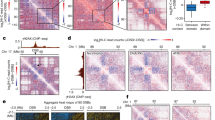
Abstract
Chromatin folds into dynamic loops that often span hundreds of kilobases and physically wire distant loci together for gene regulation. These loops are continuously created, extended and positioned by structural maintenance of chromosomes (SMC) protein complexes, such as condensin and cohesin, and their regulators, including CTCF, in a highly dynamic process known as loop extrusion. Genetic loss of extrusion factors is lethal, complicating their study. Inducible protein degradation technologies enable the depletion of loop extrusion factors within hours, leading to the rapid reconfiguration of chromatin folding. Here, we review how these technologies have changed our understanding of genome organization, upsetting long-held beliefs on its role in transcription. Finally, we examine recent models that attempt to reconcile observations after chronic versus acute perturbations, and discuss future developments in this rapidly developing field of research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Hsieh, T.-H. S. et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018). In this work, microscopy and DNA fluorescence in situ hybridization experiments were used to show that TAD structures are heterogeneous at the single-cell (and chromosome) level, and that cohesin degradation randomizes chromosome folding.
Boettiger, A. N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Misteli, T. The self-organizing genome: principles of genome architecture and function. Cell 183, 28–45 (2020).
Razin, S. V. & Ulianov, S. V. Gene functioning and storage within a folded genome. Cell. Mol. Biol. Lett. 22, 18 (2017).
Rowley, M. J. & Corces, V. G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 (2018).
Gibcus, J. H. et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). This work charts 3D genome structures leading up to pro-metaphase at high temporal resolution and after degradation of condensins resulting in a model for the formation of mitotic chromosomes.
Cremer, T. et al. Chromosome territories — a functional nuclear landscape. Curr. Opin. Cell Biol. 18, 307–316 (2006).
Hildebrand, E. M. & Dekker, J. Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci. 45, 385–396 (2020).
Solovei, I., Thanisch, K. & Feodorova, Y. How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol. 40, 47–59 (2016).
Bhat, P., Honson, D. & Guttman, M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 22, 653–670 (2021).
Ferrai, C., Castro, I. J., de, Lavitas, L., Chotalia, M. & Pombo, A. Gene positioning. Cold Spring Harb. Perspect. Biol. 2, a000588 (2010).
Hoskins, V. E., Smith, K. & Reddy, K. L. The shifting shape of genomes: dynamics of heterochromatin interactions at the nuclear lamina. Curr. Opin. Genet. Dev. 67, 163–173 (2021).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Davidson, I. F. & Peters, J.-M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 22, 445–464 (2021).
Kim, Y. & Yu, H. Shaping of the 3D genome by the ATPase machine cohesin. Exp. Mol. Med. 52, 1891–1897 (2020).
Mirny, L. A., Imakaev, M. & Abdennur, N. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 58, 142–152 (2019).
van Ruiten, M. S. & Rowland, B. D. On the choreography of genome folding: a grand pas de deux of cohesin and CTCF. Curr. Opin. Cell Biol. 70, 84–90 (2021).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–E6465 (2015).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Gomes, N. P. & Espinosa, J. M. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF–cohesin binding. Genes Dev. 24, 1022–1034 (2010).
Fudenberg, G. & Nora, E. P. Embryogenesis without CTCF in flies and vertebrates. Nat. Struct. Mol. Biol. 28, 774–776 (2021).
Singh, V. P. & Gerton, J. L. Cohesin and human disease: lessons from mouse models. Curr. Opin. Cell Biol. 37, 9–17 (2015).
Michaelis, C., Ciosk, R. & Nasmyth, K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45 (1997).
Busslinger, G. A. et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017).
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Sofueva, S. et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32, 3119–3129 (2013).
Calderon, L. et al. Cohesin-dependence of neuronal gene expression relates to chromatin loop length. eLife 11, e76539 (2022).
Yatskevich, S., Rhodes, J. & Nasmyth, K. Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019).
Nasmyth, K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001).
Pradhan, B. et al. The Smc5/6 complex is a DNA loop extruding motor. Preprint at bioRxiv https://doi.org/10.1101/2022.05.13.491800 (2022).
Venegas, A. B., Natsume, T., Kanemaki, M. & Hickson, I. D. Inducible degradation of the human SMC5/6 complex reveals an essential role only during interphase. Cell Rep. 31, 107533 (2020).
Zabolotnaya, E., Mela, I., Henderson, R. M. & Robinson, N. P. Turning the Mre11/Rad50 DNA repair complex on its head: lessons from SMC protein hinges, dynamic coiled-coil movements and DNA loop-extrusion? Biochem. Soc. Trans. 48, 2359–2376 (2020).
Gurzau, A. D., Blewitt, M. E., Czabotar, P. E., Murphy, J. M. & Birkinshaw, R. W. Relating SMCHD1 structure to its function in epigenetic silencing. Biochem. Soc. Trans. 48, 1751–1763 (2020).
Waizenegger, I. C., Hauf, S., Meinke, A. & Peters, J.-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 (2000).
Goloborodko, A., Imakaev, M. V., Marko, J. F. & Mirny, L. Compaction and segregation of sister chromatids via active loop extrusion. eLife 5, e14864 (2016).
Houlard, M. et al. MCPH1 inhibits condensin II during interphase by regulating its SMC2–kleisin interface. eLife 10, e73348 (2021).
Oldenkamp, R. & Rowland, B. D. A walk through the SMC cycle: from catching DNAs to shaping the genome. Mol. Cell 82, 1616–1630 (2022).
Elbatsh, A. M. O. et al. Distinct roles for condensin’s two ATPase sites in chromosome condensation. Mol. Cell 76, 724–737.e5 (2019).
Perea-Resa, C., Wattendorf, L., Marzouk, S. & Blower, M. D. Cohesin: behind dynamic genome topology and gene expression reprogramming. Trends Cell Biol. 31, 760–773 (2021).
Petela, N. J. et al. Scc2 is a potent activator of cohesin’s ATPase that promotes loading by binding Scc1 without Pds5. Mol. Cell 70, 1134–1148.e7 (2018).
Kikuchi, S., Borek, D. M., Otwinowski, Z., Tomchick, D. R. & Yu, H. Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc. Natl Acad. Sci. USA 113, 12444–12449 (2016).
Dauban, L. et al. Regulation of cohesin-mediated chromosome folding by Eco1 and other partners. Mol. Cell 77, 1279–1293.e4 (2020).
Wutz, G. et al. ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 9, e52091 (2020). This work shows that Hi-C after acute depletion of cohesin, WAPL/PDS5 and CTCF contrasts their relative contribution to genome folding.
van Ruiten, M. S. et al. The cohesin acetylation cycle controls chromatin loop length through a PDS5A brake mechanism. Nat. Struct. Mol. Biol. 29, 586–591 (2022).
Bastié, N. et al. Smc3 acetylation, Pds5 and Scc2 control the translocase activity that establishes cohesin-dependent chromatin loops. Nat. Struct. Mol. Biol. 29, 575–585 (2022).
van Ruiten, M. S. & Rowland, B. D. SMC complexes: universal DNA looping machines with distinct regulators. Trend Genet. 34, 477–487 (2018).
Haarhuis, J. H. I. et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707.e14 (2017).
Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
Guo, Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Vietri Rudan, M. et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015).
Li, Y. et al. The structural basis for cohesin–CTCF-anchored loops. Nature 578, 472–476 (2020).
Nora, E. P. et al. Molecular basis of CTCF binding polarity in genome folding. Nat. Commun. 11, 5612 (2020). This paper performs separation of function analysis of different protein domains by complementing inducible degradation of endogenous CTCF with mutants to show the importance of the N-terminal domain in CTCF-anchored loop formation.
Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019).
Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019).
Gabriele, M. et al. Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science 376, 496–501 (2022).
Mach, P. et al. Live-cell imaging and physical modeling reveal control of chromosome folding dynamics by cohesin and CTCF. Preprint at bioRxiv https://doi.org/10.1101/2022.03.03.482826 (2022).
Trauth, J., Scheffer, J., Hasenjäger, S. & Taxis, C. Synthetic control of protein degradation during cell proliferation and developmental processes. ACS Omega 4, 2766–2778 (2019).
Verma, R., Mohl, D. & Deshaies, R. J. Harnessing the power of proteolysis for targeted protein inactivation. Mol. Cell 77, 446–460 (2020).
Kanemaki, M. T. Ligand-induced degrons for studying nuclear functions. Curr. Opin. Cell Biol. 74, 29–36 (2022).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). This paper describes the first use of the AID system for targeted protein depletion outside plants.
Yesbolatova, A. et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11, 5701 (2020).
Li, S., Prasanna, X., Salo, V. T., Vattulainen, I. & Ikonen, E. An efficient auxin-inducible degron system with low basal degradation in human cells. Nat. Methods 16, 866–869 (2019).
Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 (2018). This paper establishes the FKBP12/dTAG system.
Nabet, B. et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 11, 4687 (2020).
Tovell, H. et al. Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader. ACS Chem. Biol. 14, 882–892 (2019).
Bond, A. G. et al. Development of BromoTag: a ‘bump-and-hole’-PROTAC system to induce potent, rapid, and selective degradation of tagged target proteins. J. Med. Chem. 64, 15477–15502 (2021).
Nowak, R. P. et al. Structure-guided design of a ‘bump-and-hole’ bromodomain-based degradation tag. J. Med. Chem. 64, 11637–11650 (2021).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Röth, S., Fulcher, L. J. & Sapkota, G. P. Advances in targeted degradation of endogenous proteins. Cell Mol. Life Sci. 76, 2761–2777 (2019).
Usherenko, S. et al. Photo-sensitive degron variants for tuning protein stability by light. BMC Syst. Biol. 8, 128 (2014).
Langan, R. A. et al. De novo design of bioactive protein switches. Nature 572, 205–210 (2019).
Samejima, K. et al. Functional analysis after rapid degradation of condensins and 3D-EM reveals chromatin volume is uncoupled from chromosome architecture in mitosis. J. Cell Sci. 131, jcs210187 (2018).
Hoencamp, C. et al. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372, 984–989 (2021).
Strom, A. R. et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 10, e63972 (2021).
Meijering, A. E. C. et al. Nonlinear mechanics of human mitotic chromosomes. Nature 605, 545–550 (2022).
Takagi, M. et al. Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. J. Cell Sci. 131, jcs212092 (2018).
Seitan, V. C. et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 23, 2066–2077 (2013).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). This work shows that acute depletion of cohesin reveals the role of cohesin in TAD and loop formation genome-wide. Restoration of cohesin following depletion shows that structures are reformed within a few hours, highlighting the dynamics of the system.
Thiecke, M. J. et al. Cohesin-dependent and -independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep. 32, 107929 (2020).
Rhodes, J. D. P. et al. Cohesin disrupts Polycomb-dependent chromosome interactions in embryonic stem cells. Cell Rep. 30, 820–835.e10 (2020).
Luppino, J. M. et al. Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat. Genet. 52, 840–848 (2020).
Szabo, Q. et al. Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat. Genet. 52, 1151–1157 (2020).
Hafner, A., Park, M., Berger, S. E., Nora, E. P. & Boettiger, A. N. Beyond TADs: microscopy reveals 2 axes of chromosome organization by CTCF and cohesin. Preprint at bioRxiv https://doi.org/10.1101/2022.07.13.499982 (2022).
Hsieh, T.-H. S. et al. Enhancer–promoter interactions and transcription are maintained upon acute loss of CTCF, cohesin, WAPL, and YY1. Preprint at bioRxiv https://doi.org/10.1101/2021.07.14.452365 (2021). This preprint provides preliminary evidence that acute cohesin of CTCF, cohesin, WAPL or YY1 only has a minor immediate effect on enhancer–promoter interactions as detected by Micro-C.
Aljahani, A. et al. Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF. Nat. Commun. 13, 2139 (2022).
Goel, V. Y., Huseyin, M. K. & Hansen, A. S. Region capture Micro-C reveals coalescence of enhancers and promoters into nested microcompartments. Preprint at bioRxiv https://doi.org/10.1101/2022.07.12.499637 (2022).
Joshi, O. et al. Dynamic reorganization of extremely long-range promoter–promoter interactions between two states of pluripotency. Cell Stem Cell 17, 748–757 (2015).
Nuebler, J., Fudenberg, G., Imakaev, M., Abdennur, N. & Mirny, L. A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl Acad. Sci. USA 115, E6697–E6706 (2018).
Zhu, Y., Denholtz, M., Lu, H. & Murre, C. Calcium signaling instructs NIPBL recruitment at active enhancers and promoters via distinct mechanisms to reconstruct genome compartmentalization. Genes Dev. 35, 65–81 (2021).
Kriz, A. J., Colognori, D., Sunwoo, H., Nabet, B. & Lee, J. T. Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb-mediated repression, and X-inactivation. Mol. Cell 81, 1970–1987.e9 (2021).
Liu, N. Q. et al. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat. Genet. 53, 100–109 (2021). This study reveals the crucial role of the cohesin loading cycle for maintaining lineage-specific gene expression.
Liu, N. Q. et al. Rapid depletion of CTCF and cohesin proteins reveals dynamic features of chromosome architecture. Preprint at bioRxiv https://doi.org/10.1101/2021.08.27.457977 (2021).
Miron, E. et al. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv. 6, eaba8811 (2020).
van Schaik, T. et al. CTCF and cohesin promote focal detachment of DNA from the nuclear lamina. Genome Biol. 23, 185 (2022).
Ladurner, R. et al. Sororin actively maintains sister chromatid cohesion. EMBO J. 35, 635–653 (2016).
Mitter, M. et al. Conformation of sister chromatids in the replicated human genome. Nature 586, 139–144 (2020).
Casa, V. et al. Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 30, 515–527 (2020).
Cuadrado, A. et al. Specific contributions of cohesin–SA1 and cohesin–SA2 to TADs and polycomb domains in embryonic stem cells. Cell Rep. 27, 3500–3510.e4 (2019).
Kojic, A. et al. Distinct roles of cohesin–SA1 and cohesin–SA2 in 3D chromosome organization. Nat. Struct. Mol. Biol. 25, 496–504 (2018).
van der Weide, R. H. et al. Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom. Bioinform 3, lqab040 (2021).
Romero-Pérez, L., Surdez, D., Brunet, E., Delattre, O. & Grünewald, T. G. P. STAG mutations in cancer. Trends Cancer 5, 506–520 (2019).
Zuin, J. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl Acad. Sci. USA 111, 996–1001 (2014).
Splinter, E. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20, 2349–2354 (2006).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). Hi-C following acute depletion of CTCF shows that CTCF acts as a boundary factor genome-wide.
Nishana, M. et al. Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation. Genome Biol. 21, 108 (2020).
Pugacheva, E. M. et al. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl Acad. Sci. USA 117, 2020–2031 (2020).
Saldana-Meyer, R. et al. RNA interactions with CTCF are essential for its proper function. Mol. Cell 76, 412–422. (2019).
Hansen, A. S. et al. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell 76, 395–411.e13 (2019).
Luan, J. et al. CTCF blocks anti-sense transcription initiation at divergent gene promoters. Preprint at bioRxiv https://doi.org/10.1101/2021.10.30.465508 (2021).
Linares-Saldana, R. et al. BRD4 orchestrates genome folding to promote neural crest differentiation. Nat. Genet. 53, 1480–1492 (2021).
Crump, N. T. et al. BET inhibition disrupts transcription but retains enhancer–promoter contact. Nat. Commun. 12, 223 (2021).
El Khattabi, L. et al. A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158.e20 (2019).
Ramasamy, S. et al. The Mediator complex regulates enhancer–promoter interactions. Preprint at bioRxiv https://doi.org/10.1101/2022.06.15.496245 (2022).
Haarhuis, J. H. I. et al. A Mediator–cohesin axis controls heterochromatin domain formation. Nat. Commun. 13, 754 (2022).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Spracklin, G. et al. Heterochromatin diversity modulates genome compartmentalization and loop extrusion barriers. Preprint at bioRxiv https://doi.org/10.1101/2021.08.05.455340 (2021).
Dequeker, B. J. H. et al. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. Nature 606, 197–203 (2022).
Banigan, E. J. et al. Transcription shapes 3D chromatin organization by interacting with loop-extruding cohesin complexes. Preprint at bioRxiv https://doi.org/10.1101/2022.01.07.475367 (2022).
Zhang, S., Übelmesser, N., Barbieri, M. & Papantonis, A. Enhancer–promoter contact formation requires RNAPII and antagonizes loop extrusion. Preprint at bioRxiv https://doi.org/10.1101/2022.07.04.498738 (2022).
Jiang, Y. et al. Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol. 21, 158 (2020).
Zhang, S. et al. RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci. Adv. 7, eabg8205 (2021). Together with Jiang et al. (2020), this work uses RNA polymerase degrons to show the limited role of these complexes in 3D genome organization, as assayed by Hi-C.
Neguembor, M. V. et al. Transcription-mediated supercoiling regulates genome folding and loop formation. Mol. Cell 81, 3065–3081.e12 (2021).
Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. & Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32, 623–626 (2002).
Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002).
Kubo, N. et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 28, 152–161 (2021).
Ottema, S. et al. The leukemic oncogene EVI1 hijacks a MYC super-enhancer by CTCF-facilitated loops. Nat. Commun. 12, 5679 (2021).
Schuijers, J. et al. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 23, 349–360 (2018).
Benabdallah, N. S. et al. Decreased enhancer–promoter proximity accompanying enhancer activation. Mol. Cell 76, 473–484.e7 (2019).
Karr, J. P., Ferrie, J. J., Tjian, R. & Darzacq, X. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 36, 7–16 (2022).
Li, J. & Pertsinidis, A. New insights into promoter–enhancer communication mechanisms revealed by dynamic single-molecule imaging. Biochem. Soc. Trans. 49, 1299–1309 (2021).
Stelloh, C. et al. The cohesin-associated protein Wapal is required for proper Polycomb-mediated gene silencing. Epigenetics Chromatin 9, 14 (2016).
Luppino, J. M. et al. NIPBL and WAPL balance cohesin activity to regulate chromatin folding and gene expression. Preprint at bioRxiv https://doi.org/10.1101/2022.04.19.488785 (2022).
Kean, C. M. et al. Decreasing Wapl dosage partially corrects transcriptome phenotypes in Nipbl–/+ embryonic mouse brain. Preprint at bioRxiv https://doi.org/10.1101/2022.05.31.493745 (2022).
Zuin, J. et al. Nonlinear control of transcription through enhancer-promoter interactions. Nature 604, 571–577 (2022).
Xie, L. et al. BRD2 compartmentalizes the accessible genome. Nat. Genet. 54, 481–491 (2022).
Rinaldi, L. et al. The glucocorticoid receptor associates with the cohesin loader NIPBL to promote long-range gene regulation. Sci. Adv. 8, eabj8360 (2022).
Gu, B. et al. Opposing effects of cohesin and transcription on CTCF organization revealed by super-resolution imaging. Mol. Cell 80, 699–711.e7 (2020).
Lee, R. et al. CTCF-mediated chromatin looping provides a topological framework for the formation of phase-separated transcriptional condensates. Nucleic Acids Res. 50, 207–226 (2022).
Nozaki, T. et al. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67, 282–293.e7 (2017).
Zuin, J. et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature 604, 571–577 (2022).
Rinzema, N. J. et al. Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat. Struct. Mol. Biol. 29, 563–574 (2022).
Kane, L. et al. Cohesin is required for long-range enhancer action. Preprint at bioRxiv https://doi.org/10.1101/2021.06.24.449812 (2021).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer–promoter loops. Cell 171, 1573–1588.e28 (2017).
Dobrinić, P., Szczurek, A. T. & Klose, R. J. PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Nat. Struct. Mol. Biol. 28, 811–824 (2021).
Barshad, G. et al. RNA polymerase II and PARP1 shape enhancer-promoter contacts. Preprint at bioRxiv https://doi.org/10.1101/2022.07.07.499190 (2022).
Hsieh, T.-H. S. et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553.e8 (2020).
Saldaña-Meyer, R. et al. RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell 76, 412–422.e5 (2019).
Stik, G. et al. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat. Genet. 52, 655–661 (2020).
Pezic, D. et al. The cohesin regulator Stag1 promotes cell plasticity through heterochromatin regulation. Preprint at bioRxiv https://doi.org/10.1101/2021.02.14.429938 (2021).
Zhang, K. et al. Analysis of genome architecture during SCNT reveals a role of cohesin in impeding minor ZGA. Mol. Cell 79, 234–250.e9 (2020).
Zhu, Y. et al. Relaxed 3D genome conformation facilitates the pluripotent to totipotent-like state transition in embryonic stem cells. Nucleic Acids Res. 49, 12167–12177 (2021).
Olbrich, T. et al. CTCF is a barrier for 2C-like reprogramming. Nat. Commun. 12, 4856 (2021).
Flavahan, W. A. et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 575, 229–233 (2019).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene–enhancer interactions. Cell 161, 1012–1025 (2015).
Valton, A.-L. & Dekker, J. TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev. 36, 34–40 (2016).
Xiao, J. Y., Hafner, A. & Boettiger, A. N. How subtle changes in 3D structure can create large changes in transcription. eLife 10, e64320 (2021).
Alexander, J. M. et al. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8, e41769 (2019).
Jerkovic, I. & Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 22, 511–528 (2021).
Kline, A. D. et al. Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat. Rev. Genet. 19, 649–666 (2018).
Solomon, D. A., Kim, J.-S. & Waldman, T. Cohesin gene mutations in tumorigenesis: from discovery to clinical significance. BMB Rep. 47, 299–310 (2014).
Kiefer, L. et al. Cohesin erases genomic-proximity biases to drive stochastic protocadherin expression for proper neural wiring. Preprint at bioRxiv https://doi.org/10.1101/2022.03.09.483674 (2022).
Canzio, D. et al. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell 177, 639–653.e15 (2019).
Ba, Z. et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586, 305–331 (2020).
Vian, L. et al. The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178.e20 (2018).
Zhang, X. et al. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature 575, 385–389 (2019).
Zhang, Y. et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019).
Dai, H.-Q. et al. Loop extrusion mediates physiological Igh locus contraction for RAG scanning. Nature 590, 338–343 (2021).
Hill, L. et al. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584, 142–147 (2020).
Arnould, C. et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 (2021).
Yunusova, A. et al. Evaluation of the OsTIR1 and AtAFB2 AID systems for genome architectural protein degradation in mammalian cells. Front. Mol. Biosci. 8, 757394 (2021).
Naqvi, S. et al. Precise modulation of transcription factor levels reveals drivers of dosage sensitivity. Preprint at bioRxiv https://doi.org/10.1101/2022.06.13.495964 (2022).
Zhang, H. et al. CTCF and transcription influence chromatin structure re-configuration after mitosis. Nat. Commun. 12, 5157 (2021).
Capece, M. et al. A novel auxin-inducible degron system for rapid, cell cycle-specific targeted proteolysis. Preprint at bioRxiv https://doi.org/10.1101/2021.04.23.441203 (2021).
Pryzhkova, M. V., Xu, M. J. & Jordan, P. W. Adaptation of the AID system for stem cell and transgenic mouse research. Stem Cell Res. 49, 102078 (2020).
Macdonald, L. et al. Rapid and specific degradation of endogenous proteins in mouse models using auxin-inducible degrons. eLife 11, e77987 (2022).
Zhang, L., Ward, J. D., Cheng, Z. & Dernburg, A. F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 (2015).
Abuhashem, A., Lee, A. S., Joyner, A. L. & Hadjantonakis, A.-K. Rapid and efficient degradation of endogenous proteins in vivo identifies stage-specific roles of RNA Pol II pausing in mammalian development. Dev. Cell 57, 1068–1080.e6 (2022).
Luan, J. et al. Distinct properties and functions of CTCF revealed by a rapidly inducible degron system. Cell Rep. 34, 108783 (2021).
Acknowledgements
The authors are grateful to L. Braccioli and R. Shah for feedback on the manuscript. E.d.W. is supported by a European Research Council (ERC) Consolidator grant FuncDis3D (865459) and is part of Oncode which is partly financed by the Dutch Cancer Society. E.P.N. is supported by National Institutes of Health (NIH) grant 1R35GM142792-01.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- CTCF
-
An 11 zinc finger DNA and RNA-binding protein able to block the extrusion of cohesin in part through its amino-terminal domain that interacts with the STAG and RAD21 cohesin subunits, thereby shielding it from WAPL-mediated release.
- Topologically associating domains
-
(TADs). Genomic regions that show preferential self-interaction and are physically insulated from their neighbours by the binding of CTCF at boundaries. Self-interaction is mediated by the cohesin complex.
- Super-enhancers
-
Multi-kilobase stretches of regulatory DNA that exhibit unusually strong occupancy of transcription factors and co-factors.
- Polycomb
-
A family of chromatin proteins that assemble into the (generally) repressive complexes Polycomb repressive complex 1 (PRC1) and PRC2, which maintain epigenetic silencing of developmental genes.
- Cohesive cohesin
-
A pool of cohesin complex that mediates sister chromatin cohesion, which depends on structural maintenance of chromosomes 3 (SMC3) acetylation, PDS5 and SORORIN binding. Cohesive cohesin is thought to be distinct from extrusive cohesin as it contains PDS5 (and not NIPBL).
- Phase separation
-
A process through which polymer chains (or segments of a polymer chain) spontaneously de-mix and segregate through the formation of immiscible phases.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Wit, E., Nora, E.P. New insights into genome folding by loop extrusion from inducible degron technologies. Nat Rev Genet 24, 73–85 (2023). https://doi.org/10.1038/s41576-022-00530-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00530-4
This article is cited by
-
Context-dependent perturbations in chromatin folding and the transcriptome by cohesin and related factors
Nature Communications (2023)