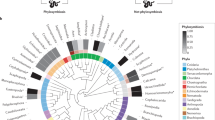
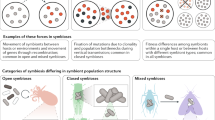
Abstract
Research on animal–microbiota interactions has become a central topic in biological sciences because of its relevance to basic eco-evolutionary processes and applied questions in agriculture and health. However, animal hosts and their associated microbial communities are still seldom studied in a systemic fashion. Hologenomics, the integrated study of the genetic features of a eukaryotic host alongside that of its associated microbes, is becoming a feasible — yet still underexploited — approach that overcomes this limitation. Acknowledging the biological and genetic properties of both hosts and microbes, along with the advantages and disadvantages of implemented techniques, is essential for designing optimal studies that enable some of the major questions in biology to be addressed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
McFall-Ngai, M. et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236 (2013).
Bosch, T. C. G. & Miller, D. J. The Holobiont Imperative: Perspective from Early Emerging Animals (Springer-Verlag, 2016).
Müller, D. B., Vogel, C., Bai, Y. & Vorholt, J. A. The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234 (2016).
Baedke, J., Fábregas-Tejeda, A. & Nieves Delgado, A. The holobiont concept before Margulis. J. Exp. Zool. B Mol. Dev. Evol. 334, 149–155 (2020).
Nyholm, S. V. & McFall-Ngai, M. The winnowing: establishing the squid–vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642 (2004).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 (2008).
Sasse, J., Martinoia, E. & Northen, T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant. Sci. 23, 25–41 (2018).
Małyska, A., Markakis, M. N., Pereira, C. F. & Cornelissen, M. The microbiome: a life science opportunity for our society and our planet. Trends Biotechnol. 37, 1269–1272 (2019).
Sessitsch, A. & Mitter, B. 21st century agriculture: integration of plant microbiomes for improved crop production and food security. Microb. Biotechnol. 8, 32 (2015).
Leidy, J. Parasites of the Termites (Collins, Printer, 1881).
Escherich, T. The intestinal bacteria of the neonate and breast-fed infant. Rev. Infect. Dis. 10, 1220–1225 (1988).
Beckwith, T. D. & Rose, E. J. Cellulose digestion by organisms from the termite gut. Proc. Soc. Exp. Biol. Med. 27, 4–6 (1929).
Margolin, S. Methods for the cultivation of cattle ciliates. Biol. Bull. 59, 301–305 (1930).
Bergeim, O., Hanszen, A. & Arnold, L. The influence of fruit ingestion before meals upon the bacterial flora of stomach and large intestine and on food allergins. Am. J. Dig. Dis. Nutr. 3, 45–52 (1936).
Lewin, H. A. et al. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl Acad. Sci. USA 115, 4325–4333 (2018).
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017).
Jefferson, R. The Hologenome. Agriculture, Environment and the Developing World: A Future of PCR (Cold Spring Harbor Press, 1994).
Nyholm, L. et al. Holo-omics: integrated host-microbiota multi-omics for basic and applied biological research. iScience 23, 101414 (2020). This article provides an overview of possible applications of hologenomics across basic and applied biological sciences.
Limborg, M. T. et al. Applied hologenomics: feasibility and potential in aquaculture. Trends Biotechnol. 36, 252–264 (2018).
Zepeda Mendoza, M. L. et al. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol. 2, 659–668 (2018).
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017).
Theis, K. R. et al. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1, e00028-16 (2016).
Moran, N. A. & Sloan, D. B. The hologenome concept: helpful or hollow? PLoS Biol. 13, e1002311 (2015).
Lloyd, E. A. & Wade, M. J. Criteria for holobionts from community genetics. Biol. Theory 14, 151–170 (2019).
Srinivas, G. et al. Genome-wide mapping of gene–microbiota interactions in susceptibility to autoimmune skin blistering. Nat. Commun. 4, 2462 (2013).
Bashiardes, S., Godneva, A., Elinav, E. & Segal, E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol. 51, 57–63 (2018).
Bermudez-Brito, M., Plaza-Díaz, J., Fontana, L., Muñoz-Quezada, S. & Gil, A. In vitro cell and tissue models for studying host–microbe interactions: a review. Br. J. Nutr. 109, S27–S34 (2013).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 83 (2017).
Argelaguet, R. et al. Multi-Omics Factor Analysis — a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018).
Singh, A. et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35, 3055–3062 (2019).
Almeida, A. et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114 (2021).
Xu, L. et al. Holo-omics for deciphering plant-microbiome interactions. Microbiome 9, 69 (2021).
Lynch, M. & Walsh, B. The Origins of Genome Architecture. Vol. 98 (Sinauer Associates, 2007).
Donoghue, P. C. J. & Purnell, M. A. Genome duplication, extinction and vertebrate evolution. Trends Ecol. Evol. 20, 312–319 (2005).
De Bodt, S., Maere, S. & Van de Peer, Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597 (2005).
Liedtke, H. C., Gower, D. J., Wilkinson, M. & Gomez-Mestre, I. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat. Ecol. Evol. 2, 1792–1799 (2018).
Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Turnbaugh, P. J., Bäckhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Peiffer, J. A. et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl Acad. Sci. USA 110, 6548–6553 (2013).
Fietz, K. et al. Mind the gut: genomic insights to population divergence and gut microbial composition of two marine keystone species. Microbiome 6, 82 (2018).
Ofek-Lalzar, M. et al. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 5, 4950 (2014).
Sumigray, K. D., Terwilliger, M. & Lechler, T. Morphogenesis and compartmentalization of the intestinal crypt. Dev. Cell 45, 183–197.e5 (2018).
Pabst, O. & Slack, E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 13, 12–21 (2020).
Bolnick, D. I. et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat. Commun. 5, 4500 (2014).
Karp, N. A. et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat. Commun. 8, 15475 (2017).
Dahlman, S., Avellaneda-Franco, L. & Barr, J. J. Phages to shape the gut microbiota? Curr. Opin. Biotechnol. 68, 89–95 (2020).
Edwards, J. A. et al. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 16, e2003862 (2018).
Danczak, R. E. et al. Members of the candidate phyla radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5, 112 (2017).
Siriyappagouder, P. et al. Exposure to yeast shapes the intestinal bacterial community assembly in zebrafish larvae. Front. Microbiol. 9, 1868 (2018).
Chabé, M., Lokmer, A. & Ségurel, L. Gut protozoa: friends or foes of the human gut microbiota? Trends Parasitol. 33, 925–934 (2017).
Leung, J. M., Graham, A. L. & Knowles, S. C. L. Parasite-microbiota interactions with the vertebrate gut: synthesis through an ecological lens. Front. Microbiol. 9, 843 (2018).
Moya, A. & Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24, 402–413 (2016).
Stanton-Geddes, J. et al. Thermal reactionomes reveal divergent responses to thermal extremes in warm and cool-climate ant species. BMC Genomics 17, 171 (2016).
Megha, S., Basu, U. & Kav, N. N. V. Regulation of low temperature stress in plants by microRNAs. Plant. Cell Env. 41, 1–15 (2018).
Yu, T. & Chen, Y. Effects of elevated carbon dioxide on environmental microbes and its mechanisms: a review. Sci. Total. Environ. 655, 865–879 (2019).
Mushegian, A. A., Arbore, R., Walser, J.-C. & Ebert, D. Environmental sources of bacteria and genetic variation in behavior influence host-associated microbiota. Appl. Environ. Microbiol. 85, e01547–18 (2019).
Vannier, N. et al. A microorganisms’ journey between plant generations. Microbiome 6, 79 (2018).
Sommer, F. et al. The gut microbiota modulates energy metabolism in the hibernating brown bear ursus arctos. Cell Rep. 14, 1655–1661 (2016).
Fung, T. C. et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 4, 2064–2073 (2019).
Li, Y. et al. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5−/− mice. Nat. Commun. 10, 1492 (2019).
Bein, A. et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol. Gastroenterol. Hepatol. 5, 659–668 (2018).
Knowles, S. C. L., Eccles, R. M. & Baltrūnaitė, L. Species identity dominates over environment in shaping the microbiota of small mammals. Ecol. Lett. 22, 826–837 (2019).
Brito, I. L. et al. Transmission of human-associated microbiota along family and social networks. Nat. Microbiol. 4, 964–971 (2019).
Michalak, L. et al. Microbiota-directed fibre activates both targeted and secondary metabolic shifts in the distal gut. Nat. Commun. 11, 5773 (2020).
Muller, P. A. et al. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature 583, 441–446 (2020). This paper shows that the microbiota modulates the expression of the neuronal transcription factor cFos through SCFAs in the gut sympathetic ganglia.
Khoruts, A. & Sadowsky, M. J. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 13, 508–516 (2016).
Bojanova, D. P. & Bordenstein, S. R. Fecal transplants: what is being transferred? PLoS Biol. 14, e1002503 (2016).
Rasmussen, T. S. et al. Bacteriophage-mediated manipulation of the gut microbiome - promises and presents limitations. FEMS Microbiol. Rev. 44, 507–521 (2020).
Vázquez-Castellanos, J. F., Biclot, A., Vrancken, G., Huys, G. R. B. & Raes, J. Design of synthetic microbial consortia for gut microbiota modulation. Curr. Opin. Pharmacol. 49, 52–59 (2019).
Kehe, J. et al. Massively parallel screening of synthetic microbial communities. Proc. Natl Acad. Sci. USA 116, 12804–12809 (2019).
Kim, H. J., Li, H., Collins, J. J. & Ingber, D. E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl Acad. Sci. USA 113, E7–E15 (2016).
D’hoe, K. et al. Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community. Elife 8, e37090 (2018).
Lawson, C. E. et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741 (2019).
Mimee, M. et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 (2018).
Riglar, D. T. & Silver, P. A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225 (2018).
McCarty, N. S. & Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197 (2019).
Ruiz-Rodríguez, M., Martín-Vivaldi, M., Martínez-Bueno, M. & Soler, J. J. Gut microbiota of great spotted cuckoo nestlings is a mixture of those of their foster magpie siblings and of cuckoo adults. Genes 9, 381 (2018).
Taylor, R. S. & Friesen, V. L. The role of allochrony in speciation. Mol. Ecol. 26, 3330–3342 (2017).
Wang, Q. et al. Host and microbiome multi-omics integration: applications and methodologies. Biophys. Rev. 11, 55–65 (2019).
Nielsen, R. L. et al. Data integration for prediction of weight loss in randomized controlled dietary trials. Sci. Rep. 10, 20103 (2020).
Sankaran, K. & Holmes, S. P. Multitable methods for microbiome data integration. Front. Genet. 10, 627 (2019).
Frankel-Bricker, J., Song, M. J., Benner, M. J. & Schaack, S. Variation in the microbiota associated with Daphnia magna across genotypes, populations, and temperature. Microb. Ecol. 79, 731–742 (2020).
Dirksen, P. et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14, 38 (2016).
Org, E. et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569 (2015).
Suzuki, T. A. et al. Host genetic determinants of the gut microbiota of wild mice. Mol. Ecol. 28, 3197–3207 (2019).
Groussin, M. et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 (2017).
Chiang, C. et al. The impact of structural variation on human gene expression. Nat. Genet. 49, 692–699 (2017).
Ansari, I. et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 5, 610–619 (2020). This article demonstrates that intestinal microorganisms induce epigenetic changes in regulatory features of host genes.
Woo, V. & Alenghat, T. Host-microbiota interactions: epigenomic regulation. Curr. Opin. Immunol. 44, 52–60 (2017).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Awany, D. et al. Host and microbiome genome-wide association studies: current state and challenges. Front. Genet. 9, 637 (2019).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Johnson, J. S. et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029 (2019).
Goyal, A., Bittleston, L. S., Leventhal, G. E., Lu, L. & Cordero, O. X. Interactions between strains govern the eco-evolutionary dynamics of microbial communities. bioRxiv https://doi.org/10.1101/2021.01.04.425224 (2021).
Antony-Babu, S. et al. Multiple Streptomyces species with distinct secondary metabolomes have identical 16S rRNA gene sequences. Sci. Rep. 7, 11089 (2017).
Aßhauer, K. P., Wemheuer, B., Daniel, R. & Meinicke, P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015).
Iwai, S. et al. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 11, e0166104 (2016).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
Sun, S., Jones, R. B. & Fodor, A. A. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 8, 46 (2020).
Lesker, T. R. et al. An integrated metagenome catalog reveals new insights into the murine gut microbiome. Cell Rep. 30, 2909–2922.e6 (2020).
Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021).
Overholt, W. A. et al. Inclusion of Oxford Nanopore long reads improves all microbial and viral metagenome-assembled genomes from a complex aquifer system. Environ. Microbiol. 22, 4000–4013 (2020).
Jiang, Y., Balaban, M., Zhu, Q. & Mirarab, S. DEPP: deep learning enables extending species trees using single genes. bioRxiv https://doi.org/10.1101/2021.01.22.427808 (2021).
Zhao, L. et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018).
Minot, S. S. & Willis, A. D. Clustering co-abundant genes identifies components of the gut microbiome that are reproducibly associated with colorectal cancer and inflammatory bowel disease. Microbiome 7, 110 (2019).
Martino, C. et al. Context-aware dimensionality reduction deconvolutes gut microbial community dynamics. Nat. Biotechnol. 39, 165–168 (2021).
Xu, Y. Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 129, 653–673 (2016).
Foote, A. D. et al. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11693 (2016).
Rojas, C. A., Holekamp, K. E., Winters, A. D. & Theis, K. R. Body site-specific microbiota reflect sex and age-class among wild spotted hyenas. FEMS Microbiol. Ecol. 96, fiaa007 (2020).
Riva, A. et al. A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome. Nat. Commun. 10, 4366 (2019).
Sheth, R. U. et al. Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol. 37, 877–883 (2019).
Zaborin, A. et al. Spatial compartmentalization of the microbiome between the lumen and crypts is lost in the murine cecum following the process of surgery, including overnight fasting and exposure to antibiotics. mSystems 5, e00377 (2020).
Ellegaard, K. M. & Engel, P. Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10, 446 (2019).
Shah, T. M., Patel, J. G., Gohil, T. P., Blake, D. P. & Joshi, C. G. Host transcriptome and microbiome interaction modulates physiology of full-sibs broilers with divergent feed conversion ratio. NPJ Biofilms Microb. 5, 24 (2019).
Fukami, T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46, 1–23 (2015).
Gleason, H. A. Further views on the succession-concept. Ecology 8, 299–326 (1927).
Debray, R. et al. Priority effects in microbiome assembly. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-021-00604-w (2021).
Sprockett, D., Fukami, T. & Relman, D. A. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 15, 197–205 (2018).
Martínez, I. et al. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521 (2018).
Johnson, A. J. et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25, 789–802.e5 (2019).
Smits, S. A. et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806 (2017).
Knutie, S. A., Wilkinson, C. L., Kohl, K. D. & Rohr, J. R. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 8, 86 (2017).
Arnold, I. C. et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J. Clin. Invest. 121, 3088–3093 (2011).
Lynch, J. B. & Hsiao, E. Y. Microbiomes as sources of emergent host phenotypes. Science 365, 1405–1409 (2019).
Mayoral-Peña, Z., Álvarez-Martínez, R., Fornoni, J. & Garrido, E. In Evolutionary Ecology of Plant-Herbivore Interaction (eds Núñez-Farfán, J. & Valverde, P. L.) 135–146 (Springer International Publishing, 2020).
Lindsay, E. C., Metcalfe, N. B. & Llewellyn, M. S. The potential role of the gut microbiota in shaping host energetics and metabolic rate. J. Anim. Ecol. 89, 2415–2426 (2020).
Vaelli, P. M. et al. The skin microbiome facilitates adaptive tetrodotoxin production in poisonous newts. eLife 9, e53898 (2020). This work demonstrates that the toxic newt phenotype is shaped through the combination of toxin-producing skin microorganisms and toxin-tolerant host genotypes.
Thaiss, C. A. et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12 (2016).
Nichols, R. G. & Davenport, E. R. The relationship between the gut microbiome and host gene expression: a review. Hum. Genet. 140, 747–760 (2021).
Chen, H. et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 177, 1217–1231.e18 (2019).
Yano, J. M. et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 (2015).
Meisel, J. S. et al. Commensal microbiota modulate gene expression in the skin. Microbiome 6, 20 (2018).
Rooks, M. G. & Garrett, W. S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016).
Sanchez, H. N. et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat. Commun. 11, 60 (2020). This study shows that microbiota-produced SCFAs impair intestinal and systemic antibody responses upregulating micro RNAs that target immune genes through inhibition of histone deacetylation.
Bonder, M. J. et al. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016).
Poole, A. C. et al. Human Salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 25, 553–564.e7 (2019).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Wagner, M. R. et al. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 12151 (2016).
Qanbari, S. et al. Genetics of adaptation in modern chicken. PLoS Genet. 15, e1007989 (2019).
Weissbrod, O., Rothschild, D., Barkan, E. & Segal, E. Host genetics and microbiome associations through the lens of genome wide association studies. Curr. Opin. Microbiol. 44, 9–19 (2018).
Suzuki, T. A. Links between natural variation in the microbiome and host fitness in wild mammals. Integr. Comp. Biol. 57, 756–769 (2017).
Gould, A. L. et al. Microbiome interactions shape host fitness. Proc. Natl Acad. Sci. U. S. A. 115, E11951–E11960 (2018).
Rosshart, S. P. et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1021.e13 (2017).
Cooper, R. O., Vavra, J. M. & Cressler, C. E. Targeted manipulation of abundant and rare taxa in the daphnia magna microbiota with antibiotics impacts host fitness differentially. mSystems 6, e00916-20 (2021).
Moeller, A. H. & Sanders, J. G. Roles of the gut microbiota in the adaptive evolution of mammalian species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190597 (2020).
Fontaine, S. S. & Kohl, K. D. Optimal integration between host physiology and functions of the gut microbiome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190594 (2020).
Suzuki, T. A. & Ley, R. E. The role of the microbiota in human genetic adaptation. Science 370, aaz6827 (2020).
Alberdi, A., Aizpurua, O., Bohmann, K., Zepeda-Mendoza, M. L. & Gilbert, M. T. P. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699 (2016).
Wang, G.-H. et al. Changes in microbiome confer multigenerational host resistance after sub-toxic pesticide exposure. Cell Host Microbe 27, 213–224.e7 (2020). This paper shows that the gut microbiota confers Nasonia wasps with resistance towards pesticides and that this acquired trait shapes their genomic evolutionary features across multiple generations.
van Opstal, E. J. & Bordenstein, S. R. Rethinking heritability of the microbiome. Science 349, 1172–1173 (2015).
Douglas, G. M., Bielawski, J. P. & Langille, M. G. I. Re-evaluating the relationship between missing heritability and the microbiome. Microbiome 8, 87 (2020).
Khan, A. A. et al. Polymorphic immune mechanisms regulate commensal repertoire. Cell Rep. 29, 541–550.e4 (2019). This study shows that genetic variants of innate and adaptive immunity genes shape distinct microbial communities through the production of distinct immunoglobulins some microorganisms use for colonizing the mucosal barrier.
Davenport, E. R. Genetic variation shapes murine gut microbiota via immunity. Trends Immunol. 41, 1–3 (2020).
Van Vliet, S. & Doebeli, M. The role of multilevel selection in host microbiome evolution. Proc. Natl Acad. Sci. USA 116, 20591–20597 (2019).
Groussin, M., Mazel, F. & Alm, E. J. Co-evolution and co-speciation of host-gut bacteria systems. Cell Host Microbe 28, 12–22 (2020).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Beaulaurier, J. et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat. Biotechnol. 36, 61–69 (2018).
Ma, F., Jiang, S. & Zhang, C.-Y. Recent advances in histone modification and histone modifying enzyme assays. Expert Rev. Mol. Diagn. 19, 27–36 (2019).
DeMaere, M. Z. & Darling, A. E. bin3C: exploiting Hi-C sequencing data to accurately resolve metagenome-assembled genomes. Genome Biol. 20, 46 (2019).
Kent, A. G., Vill, A. C., Shi, Q., Satlin, M. J. & Brito, I. L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379 (2020).
Hammer, T. J., Janzen, D. H., Hallwachs, W., Jaffe, S. P. & Fierer, N. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA 114, 9641–9646 (2017).
Oliver, K. M., Moran, N. A. & Hunter, M. S. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. Biol. Sci. 273, 1273–1280 (2006).
Song, S. J. et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio 11, e02901-19 (2020).
Krediet, C. J., Ritchie, K. B., Paul, V. J. & Teplitski, M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. Biol. Sci. 280, 20122328 (2013).
Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180 (2014).
McCann, J. C., Wickersham, T. A. & Loor, J. J. High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinform. Biol. Insights 8, 109–125 (2014).
Rosshart, S. P. et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, aaw4361 (2019).
Niehus, R., Mitri, S., Fletcher, A. G. & Foster, K. R. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat. Commun. 6, 8924 (2015).
Carrier, T. J. & Reitzel, A. M. The hologenome across environments and the implications of a host-associated microbial repertoire. Front. Microbiol. 8, 802 (2017).
Mushegian, A. A. & Ebert, D. Rethinking ‘mutualism’ in diverse host-symbiont communities. Bioessays 38, 100–108 (2016).
Pereira, F. C. & Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 19, 1366–1378 (2017).
Erin Chen, Y., Fischbach, M. A. & Belkaid, Y. Skin microbiota–host interactions. Nature 553, 427–436 (2018).
Earley, A. M., Graves, C. L. & Shiau, C. E. Critical role for a subset of intestinal macrophages in shaping gut microbiota in adult Zebrafish. Cell Rep. 25, 424–436 (2018).
Kartzinel, T. R., Hsing, J. C. & Musili, P. M. Covariation of diet and gut microbiome in African megafauna. Proc. Natl Acad. Sci. USA 116, 23588–23593 (2019).
Perofsky, A. C., Lewis, R. J., Abondano, L. A., Di Fiore, A. & Meyers, L. A. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proc. Biol. Sci. 284, 20172274 (2017).
Asnicar, F. et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2, e00164-16 (2017).
Storelli, G. et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414 (2011).
Johnson, K. V.-A. & Foster, K. R. Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655 (2018).
Davidson, G. L., Raulo, A. & Knowles, S. C. L. Identifying microbiome-mediated behaviour in wild vertebrates. Trends Ecol. Evol. 35, 972–980 (2020).
Zhang, Y.-J. et al. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 16, 7493–7519 (2015).
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P. & Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306 (2012).
Nicholson, J. K. et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012).
Fellows, R. et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 9, 105 (2018).
Neuman, H., Debelius, J. W., Knight, R. & Koren, O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 39, 509–521 (2015).
Acknowledgements
The authors thank the Danish National Research Foundation award DNRF143 ‘A Center for Evolutionary Hologenomics’ for funding their research. A.A. acknowledges Lundbeckfonden grant R250-2017-1351. A.A., M.T.L. and M.T.P.G. were supported by the European Union (H2020-SFS-2018-1 project HoloFood-817729). M.T.L. and M.T.P.G. acknowledge the FHF (Norwegian Seafood Research Fund; “HoloFish”, grant No. 901436). S.B.A. acknowledges Lundbeckfonden Fellowship R335-2019-1513 ‘Understanding the Health Effects of Microbial Interactions’ and Independent Research Council Denmark Sapere Aude grant 9064-00029B ‘Understanding the Effects of Microbial Interactions on Host Health’.
Author information
Authors and Affiliations
Contributions
A.A. researched data for the article. All authors made substantial contributions to discussions of the content and writing the article and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks S. Borderstein, T. Bosch and M. van Oppen for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Metagenotype
-
The specific state of the microbial metagenome characterized at a particular moment and at a given resolution.
- Hologenotype
-
The entire genetic constitution of an individual eukaryotic organism and its associated microorganisms characterized at a given moment and at a given resolution.
- Metagenome-assembled genomes
-
(MAGs). Partial or semi-complete draft bacterial genomes reconstructed through metagenomic assembly and binning from samples containing mixtures of microbial taxa.
Rights and permissions
About this article
Cite this article
Alberdi, A., Andersen, S.B., Limborg, M.T. et al. Disentangling host–microbiota complexity through hologenomics. Nat Rev Genet 23, 281–297 (2022). https://doi.org/10.1038/s41576-021-00421-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-021-00421-0
This article is cited by
-
A first characterization of the microbiota-resilience link in swine
Microbiome (2024)
-
Ecological filtering and phylogeographic structuring of Psychrilyobacter within two closely related limpet species from the Southern Ocean
Annals of Microbiology (2024)
-
Intestinal dual-specificity phosphatase 6 regulates the cold-induced gut microbiota remodeling to promote white adipose browning
npj Biofilms and Microbiomes (2024)
-
Host genetic regulation of human gut microbial structural variation
Nature (2024)
-
The gastrointestinal microbiome in dairy cattle is constrained by the deterministic driver of the region and the modified effect of diet
Microbiome (2023)