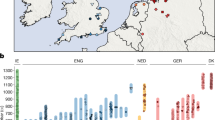
Abstract
In the field of human history, ancient DNA has provided answers to long-standing debates about major movements of people and has begun to inform on other important facets of the human experience. The field is now moving from mostly large-scale supraregional studies to a more local perspective, shedding light on socioeconomic processes, inheritance rules, marriage practices and technological diffusion. In this Review, we summarize recent studies showcasing these types of insights, focusing on methods used to infer sociocultural aspects of human behaviour. This approach often involves working across disciplines — such as anthropology, archaeology, linguistics and genetics — that have until recently evolved in separation. Multidisciplinary dialogue is important for an integrated reconstruction of human history, which can yield extraordinary insights about past societies, reproductive behaviours and even lifestyle habits that would not be possible to obtain otherwise.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ammerman, A. J. & Cavalli-Sforza, L. L. The Neolithic Transition and the Genetics of Populations in Europe (Princeton University Press, 1984)
Hakenbeck, S. Migration in archaeology: are we nearly there yet? Archaeol. Rev. Camb. 23, 9–26 (2008).
Hofreiter, M., Serre, D., Poinar, H. N., Kuch, M. & Pääbo, S. Ancient DNA. Nat. Rev. Genet. 2, 353–359 (2001).
Willerslev, E. & Cooper, A. Ancient DNA. Proc. Biol. Sci. 272, 3–16 (2005).
Damgaard, P. B. et al. Improving access to endogenous DNA in ancient bones and teeth. Sci. Rep. 5, 11184 (2015).
Pinhasi, R. et al. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10, e0129102 (2015).
Sirak, K. A. et al. A minimally-invasive method for sampling human petrous bones from the cranial base for ancient DNA analysis. Biotechniques 62, 283–289 (2017).
Cruz-Dávalos, D. I. et al. Experimental conditions improving in-solution target enrichment for ancient DNA. Mol. Ecol. Resour. 17, 508–522 (2017).
Dabney, J. & Meyer, M. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 52, 87–94 (2012).
Gansauge, M.-T. & Meyer, M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748 (2013).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012).
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012).
Barnett, R. & Larson, G. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol. Biol. 840, 13–19 (2012).
Carpenter, M. L. et al. Pulling out the 1%: whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864 (2013).
Slatkin, M. & Racimo, F. Ancient DNA and human history. Proc. Natl Acad. Sci. USA 113, 6380–6387 (2016).
Marciniak, S. & Perry, G. H. Harnessing ancient genomes to study the history of human adaptation. Nat. Rev. Genet. 18, 659–674 (2017).
Skoglund, P. & Mathieson, I. Ancient genomics of modern humans: the first decade. Annu. Rev. Genomics Hum. Genet. 19, 381–404 (2018).
Pickrell, J. & Reich, D. Towards a new history and geography of human genes informed by ancient DNA. Trends. Genet. 30, 377–389 (2014).
Gerstenberger, J., Hummel, S. & Herrmann, B. Reconstruction of residence patterns through genetic typing of skeletal remains of an early medieval population. Anc. Biomol. 4, 25–32 (2002).
Gerstenberger, J., Hummel, S., Schultes, T., Häck, B. & Herrmann, B. Reconstruction of a historical genealogy by means of STR analysis and Y-haplotyping of ancient DNA. Eur. J. Hum. Genet. 7, 469–477 (1999).
Haak, W. et al. Ancient DNA, strontium isotopes, and osteological analyses shed light on social and kinship organization of the later Stone Age. Proc. Natl Acad. Sci. USA 105, 18226–18231 (2008).
Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014).
Lindo, J. et al. A time transect of exomes from a Native American population before and after European contact. Nat. Commun. 7, 13175 (2016).
Irving-Pease, E. K. et al. Paleogenomics of animal domestication. Popul. Genomics. https://doi.org/10.1007/13836_2018_55 (2018).
Estrada, O., Breen, J., Richards, S. M. & Cooper, A. Ancient plant DNA in the genomic era. Nat. Plants 4, 394–396 (2018).
Gutaker, R. M. & Burbano, H. A. Reinforcing plant evolutionary genomics using ancient DNA. Curr. Opin. Plant. Biol. 36, 38–45 (2017).
MacHugh, D. E., Larson, G. & Orlando, L. Taming the past: ancient DNA and the study of animal domestication. Annu. Rev. Anim. Biosci. 5, 329–351 (2017).
Houldcroft, C. J., Rifkin, R. F. & Underdown, S. J. Human biology and ancient DNA: exploring disease, domestication and movement. Ann. Hum. Biol. 46, 95–98 (2019).
McHugo, G. P., Dover, M. J. & MacHugh, D. E. Unlocking the origins and biology of domestic animals using ancient DNA and paleogenomics. BMC Biol. 17, 98 (2019).
Bocquet-Appel, J.-P., Naji, S., Linden, M. V. & Kozlowski, J. K. Detection of diffusion and contact zones of early farming in Europe from the space-time distribution of 14C dates. J. Arch. Sci. 36, 807–820 (2009).
Silva, F. & Steele, J. New methods for reconstructing geographical effects on dispersal rates and routes from large-scale radiocarbon databases. J. Arch. Sci. 52, 609–620 (2014).
Fort, J. Demic and cultural diffusion propagated the Neolithic transition across different regions of Europe. J. R. Soc. Interface https://doi.org/10.1098/rsif.2015.0166 (2015).
Silva, F. & Linden, M. V. Amplitude of travelling front as inferred from 14C predicts levels of genetic admixture among European early farmers. Sci. Rep. 7, 11985 (2017).
Fort, J. in Diffusive Spreading in Nature, Technology and Society (eds Bunde, A., Caro, J., Kärger, J. & Vogl, G.) 313–331 (Springer, 2018).
Lemmen, C. & Gronenborn, D. in Diffusive Spreading in Nature, Technology and Society (eds Bunde, A., Caro, J., Kärger, J. & Vogl, G.) 333–349 (Springer, 2018)
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). The authors perform a genome-wide selection analysis in a large data set of ancient European genomes and are able to describe temporal changes in adaptive alleles. This work paved the way for future large screenings to detect selective events.
Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016).
Skoglund, P. et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466–469 (2012).
Skoglund, P. et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750 (2014).
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
Mathieson, I. et al. The genomic history of southeastern Europe. Nature 555, 197–203 (2018).
Broushaki, F. et al. Early Neolithic genomes from the eastern Fertile Crescent. Science 353, 499–503 (2016).
Racimo, F. et al. A geostatistical approach to modelling human Holocene migrations in Europe using ancient DNA. Preprint at bioRxiv https://doi.org/10.1101/826149 (2019).
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014).
Lipson, M. et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017).
Hofmanová, Z. et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl Acad. Sci. USA 113, 6886–6891 (2016).
Sikora, M. et al. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 10, e1004353 (2014).
Allentoft, M. E. et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). Haak et al. (2015) and Allentoft et al. (2015) are two key articles that independently identify signals for migration from the Eurasian Steppe in the third millennium BCE in Europe.
Narasimhan, V. M. et al. The formation of human populations in South and Central Asia. Science 365, eaat7478 (2019). This work explores the movements of people in the past thousands of years and the formation of a male-biased genetic cline from Central Asia into South Asia, which may be associated with the spread of Indo-European languages and other social transformation events.
Lipson, M. et al. Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95 (2018).
McColl, H. et al. The prehistoric peopling of Southeast Asia. Science 361, 88–92 (2018).
Prendergast, M. E. et al. Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science https://doi.org/10.1126/science.aaw6275 (2019).
Ye, K., Gao, F., Wang, D., Bar-Yosef, O. & Keinan, A. Dietary adaptation of FADS genes in Europe varied across time and geography. Nat. Ecol. Evolution 1, 0167 (2017).
Buckley, M. T. et al. Selection in Europeans on fatty acid desaturases associated with dietary changes. Mol. Biol. Evol. 34, 1307–1318 (2017).
Mathieson, S. & Mathieson, I. FADS1 and the timing of human adaptation to agriculture. Mol. Biol. Evol. 35, 2957–2970 (2018).
Bevan, A. et al. Holocene fluctuations in human population demonstrate repeated links to food production and climate. Proc. Natl Acad. Sci. USA 114, E10524–E10531 (2017).
Capuzzo, G., Zanon, M., Dal Corso, M., Kirleis, W. & Barceló, J. A. Highly diverse Bronze Age population dynamics in central-southern Europe and their response to regional climatic patterns. PLoS One 13, e0200709 (2018).
Czebreszuk, J. Similar but Different: Bell Beakers in Europe (Sidestone Press, 2014).
Fokkens, H. & Nicolis, F. Background to Beakers: Inquiries into the Regional Cultural Background to the Bell Beaker Complex. (Sidestone Press, 2012).
Shennan, S. J. Settlement and social change in central Europe, 3500-1500 BC. J. World Prehist. 7, 121–161 (1993).
Linden, M. V. What linked the Bell Beakers in third millennium BC Europe? Antiquity 81, 343–352 (2007).
Olalde, I. et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018). This continental-scale analysis on the Bell Beaker archaeological phenomenon unveiled first a diffusion of ideas and a subsequent population movement that replaced the previous Neolithic population from the British Isles.
Outram, A. K. et al. The earliest horse harnessing and milking. Science 323, 1332–1335 (2009).
de Barros Damgaard, P. et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360, eaa7711 (2018).
Gaunitz, C. et al. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 360, 111–114 (2018).
Fages, A. et al. Tracking five millennia of horse management with extensive ancient genome time series. Cell 177, 1419–1435 (2019).
Sánchez-Quinto, F. et al. Megalithic tombs in western and northern Neolithic Europe were linked to a kindred society. Proc. Natl Acad. Sci. USA 116, 9469–9474 (2019).
Scarre, C. J. Archaeology: kin-groups in Megalithic burials. Nature 311, 512–513 (1984).
Schroeder, H. et al. Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc. Natl Acad. Sci. USA 116, 10705–10710 (2019). The genetic analysis of a Late Neolithic Polish mass grave with 15 bodies exposed the kinship links among the people buried in this dramatic episode at the time of the arrival of the Eurasian Steppe nomads.
Knipper, C. et al. Female exogamy and gene pool diversification at the transition from the Final Neolithic to the Early Bronze Age in central Europe. Proc. Natl Acad. Sci. USA 114, 10083–10088 (2017).
Kennett, D. J. et al. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 8, 14115 (2017).
Margaryan, A. et al. Eight millennia of matrilineal genetic continuity in the south Caucasus. Curr. Biol. 27, 2023–2028 (2017).
Degnan, J. H. & Rosenberg, N. A. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340 (2009).
Peletz, M. G. Kinship studies in late twentieth-century anthropology. Annu. Rev. Anthropol. 24, 343–372 (1995).
Mittnik, A. et al. Kinship-based social inequality in Bronze Age Europe. Science 366, 731–734 (2019). The genetic analysis of several Bronze Age settlements in southern Germany helped to understand the patrilineal social structure underlying genetic transformation in that period.
Amorim, C. E. G. et al. Understanding 6th-century barbarian social organization and migration through paleogenomics. Nat. Commun. 9, 3547 (2018). This is another example of integration of ancient DNA and several other sources of evidence for past behaviour at an individual site.
Goldberg, A., Günther, T., Rosenberg, N. A. & Jakobsson, M. Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations. Proc. Natl Acad. Sci. USA 114, 2657–2662 (2017).
Lazaridis, I. & Reich, D. Failure to replicate a genetic signal for sex bias in the steppe migration into central Europe. Proc. Natl Acad. Sci. USA 114, E3873–E3874 (2017).
Goldberg, A., Günther, T., Rosenberg, N. A. & Jakobsson, M. Reply to Lazaridis and Reich: robust model-based inference of male-biased admixture during Bronze Age migration from the Pontic-Caspian Steppe. Proc. Natl Acad. Sci. USA 114, E3875–E3877 (2017).
Olalde, I. et al. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234 (2019). This work is an example of the large amount of information that can be generated from a single temporal transect in a specific geographical region. The researchers uncovered a male-biased population turnover during the Bronze Age in the Iberian Peninsula.
Martiniano, R. et al. The population genomics of archaeological transition in west Iberia: investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13, e1006852 (2017).
Saag, L. et al. Extensive farming in Estonia started through a sex-biased migration from the steppe. Curr. Biol. 27, 2185–2193 (2017).
Sandoval-Velasco, M. et al. The genetic origins of Saint Helena’s liberated Africans. Preprint at bioRxiv https://doi.org/10.1101/787515 (2019).
Ginzburg, C. Microhistory: two or three things that I know about it. Crit. Inq. 20, 10–35 (1993).
González-Fortes, G. et al. A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc. Biol. Sci. 286, 20182288 (2019).
Schuhmacher, T. X., Cardoso, J. L. & Banerjee, A. Sourcing African ivory in Chalcolithic Portugal. Antiquity 83, 983–997 (2009).
Margaryan, A., Lawson, D., Sikora, M. & Racimo, F. Population genomics of the Viking world. Preprint at bioRxiv https://doi.org/10.1101/703405 (2019).
Harney, É. et al. Ancient DNA from the skeletons of Roopkund Lake reveals Mediterranean migrants in India. Nat. Commun. 10, 3670 (2019).
Loog, L. et al. Estimating mobility using sparse data: application to human genetic variation. Proc. Natl Acad. Sci. USA 114, 12213–12218 (2017). This report presents a statistical method that can estimate how much mobility existed among peoples in a given region during periods for which several ancient genomes are available.
Knapp, A. B. & van Dommelen, P. Past practices: rethinking individuals and agents in archaeology. Camb. Archaeol. J. 18, 15–34 (2008).
Jensen, T. Z. T., Niemann, J., Iversen, K. H. & Fotakis, A. K. Stone Age ‘chewing gum’ yields 5,700 year-old human genome and oral microbiome. Nat. Commun. 10, 5520 (2019).
Kashuba, N. et al. Ancient DNA from mastics solidifies connection between material culture and genetics of Mesolithic hunter–gatherers in Scandinavia. Commun Biol. 2, 185 (2019). Jensen et al. (2019) and Kashuba et al. (2019). Two seminal studies demonstrating the potential of chewed birch pitch for recovery of ancient DNA.
Schablitsky, J. M. et al. Ancient DNA analysis of a nineteenth century tobacco pipe from a Maryland slave quarter. J. Archaeol. Sci. 105, 11–18 (2019).
Lalueza-Fox, C. Agreements and misunderstandings among three scientific fields. Curr. Anthropol. 54, S214–S220 (2013).
Furholt, M. Massive migrations? The impact of recent aDNA studies on our view of third millennium Europe. European. J. Archaeol. 21, 159–191 (2018).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Lake, M. W. Trends in archaeological simulation. J. Archaeol. Method Theory 21, 258–287 (2014).
Bradburd, G. S. & Ralph, P. L. Spatial population genetics: it’s about time. Annu. Rev. Ecol. Evol. Syst. 50, 427–449 (2019).
Prendergast, M. E. & Sawchuk, E. Boots on the ground in Africa’s ancient DNA ‘revolution’: archaeological perspectives on ethics and best practices. Antiquity 92, 803–815 (2018).
Fox, K. & Hawks, J. Use ancient remains more wisely. Nature 572, 581–583 (2019).
Cotterman, C. W. A Calculus for Statistico-genetics https://etd.ohiolink.edu/!etd.send_file?accession=osu1298297334&disposition=inline (Ohio State University, Columbus, 1940).
Choi, Y., Wijsman, E. M. & Weir, B. S. Case-control association testing in the presence of unknown relationships. Genet. Epidemiol. 33, 668–678 (2009).
Thompson, E. A. The estimation of pairwise relationships. Ann. Hum. Genet. 39,173–188 (1975).
Gusev, A. et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 19, 318–326 (2008).
Browning, B. L. & Browning, S. R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Albrechtsen, A., Moltke, I. & Nielsen, R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics 186, 295–308 (2010).
Korneliussen, T. S. & Moltke, I. NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31, 4009–4011 (2015).
Martin, M. D., Jay, F., Castellano, S. & Slatkin, M. Determination of genetic relatedness from low-coverage human genome sequences using pedigree simulations. Mol. Ecol. 26, 4145–4157 (2017).
Theunert, C., Racimo, F. & Slatkin, M. Joint estimation of relatedness coefficients and allele frequencies from ancient samples. Genetics 206, 1025–1035 (2017).
Kuhn, J. M. M., Jakobsson, M. & Günther, T. Estimating genetic kin relationships in prehistoric populations. PLoS One 13, e0195491 (2018).
Waples, R. K., Albrechtsen, A. & Moltke, I. Allele frequency-free inference of close familial relationships from genotypes or low-depth sequencing data. Mol. Ecol. 28, 35–48 (2019).
Thompson, E. A. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics 194, 301–326 (2013).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Narasimhan, V. et al. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32, 1749–1751 (2016).
Su, S.-Y. et al. Detection of identity by descent using next-generation whole genome sequencing data. BMC Bioinformatics 13, 121 (2012).
Bertrand, A. R., Kadri, N. K., Flori, L., Gautier, M. & Druet, T. RZooRoH: an R package to characterize individual genomic autozygosity and identify homozygous-by-descent segments. Methods Ecol. Evol. 10, 860–866 (2019).
Vieira, F. G., Albrechtsen, A. & Nielsen, R. Estimating IBD tracts from low coverage NGS data. Bioinformatics 32, 2096–2102 (2016).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Prüfer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017).
Prüfer, K. snpAD: an ancient DNA genotype caller. Bioinformatics 34, 4165–4171 (2018).
Sikora, M. et al. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science 358, 659–662 (2017). This is a seminal study inferring social and reproductive behaviour among early Late Palaeolithic foragers from ancient genomes.
Palamara, P. F., Lencz, T., Darvasi, A. & Pe’er, I. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 91, 809–822 (2012).
Linden, M. V. & Silva, F. Comparing and modeling the spread of early farming across Europe. Past. Glob. Change Mag. 26, 28–29 (2018).
Sikora, M. et al. The population history of northeastern Siberia since the Pleistocene. Nature 570, 182–188 (2019).
Browning, B. L. & Browning, S. R. Detecting identity by descent and estimating genotype error rates in sequence data. Am. J. Hum. Genet. 93, 840–851 (2013).
Wang, C.-C. et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 10, 590 (2019).
Acknowledgements
The authors thank F. Silva for providing raw data for reproduction of agricultural spread patterns featured in a previous article by him. F.R. was funded by a Villum Fonden Young Investigator award (project no. 00025300). H.S. was supported by the Humanities in the European Research Area (HERA) Joint Research Programme “Uses of the Past” (CitiGen) and the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 649307. M.V.L. was supported by the European Research Council, funded under the European Union’s Seventh Framework Programme (FP/20072013; European Research Council grant agreement no. 313716). C.L.-F. was supported by a grant from Obra Social “La Caixa” (GRC2017-SGR880, Generalitat de Catalunya) and a grant from FEDER-MCIU-AEI (PGC2018-095931-B-100) of Spain.
Author information
Authors and Affiliations
Contributions
F.R., M.S., M.V.L. and H.S. researched the literature. All authors provided substantial contributions to discussions of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks G. H. Perry, W. Haak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Population admixture
-
The introduction of genetic lineages from one population into another population that is genetically differentiated from it, because of interbreeding between them at some point in the past.
- Eurasian Steppe
-
Large region of Eurasia extending from eastern Europe through Russia to Mongolia, which served as a major transit and connection route for many cultures throughout history and prehistory; characterized by temperate grasslands, shrub lands and savannahs.
- Cultural diffusion
-
The spread of technologies or cultures from one region to another via the exchange of ideas between populations, with limited associated movements of people. By contrast, demic diffusion is the spread of technologies or cultures via movement of people, often prompted by population growth and expansion.
- Megalithic
-
Pertaining to large stone structures, tombs or monuments. In Europe, the practice of megalith construction took off mainly in the Neolithic period, reached an apogee during the Chalcolithic period and continued into the Bronze Age.
- Patrilineal
-
A term to describe a kinship system in which a person’s social status, family membership and/or property rights are determined through that person’s paternal lineage. By contrast, in a matrilineal system, these factors are determined through the maternal lineage.
- Exogamy
-
The cultural practice by which individuals tend to marry outside their immediate kin group.
- Identical by descent
-
(IBD). Two segments from two different genomes are IBD if they were both inherited from a recent ancestor shared between the two genomes.
- Homozygous by descent
-
(HBD). Genomic segments shared identical by descent within the same individual, resulting in continuous stretches of homozygous genotypes termed ‘runs of homozygosity’.
- Endogamy
-
The cultural practice by which individuals tend to marry within their own kin group or social group.
- Uniparentally inherited markers
-
Sequences of DNA that are — barring rare exceptions — inherited from only one or another of a person’s parents. Examples include the mitochondrial DNA genome (transmitted from the mother alone) and the Y chromosome genome (transmitted from fathers to sons).
- Agent-based models
-
Computational models designed for simulating and studying the behaviour of multiple autonomous agents that may interact with each other so as to study their collective effects on a system.
- Isotope analysis
-
The study of the concentrations of different varieties of a chemical element — such as carbon, nitrogen or strontium — that have different numbers of neutrons in biological samples. They can indicate the relative abundance of vegetation types or dietary items or identify non-local individuals.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Racimo, F., Sikora, M., Vander Linden, M. et al. Beyond broad strokes: sociocultural insights from the study of ancient genomes. Nat Rev Genet 21, 355–366 (2020). https://doi.org/10.1038/s41576-020-0218-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-020-0218-z
This article is cited by
-
Detection of chromosomal aneuploidy in ancient genomes
Communications Biology (2024)
-
Human DNA from a prehistoric artefact
Nature Reviews Genetics (2023)
-
Genome-wide analysis of a collective grave from Mentesh Tepe provides insight into the population structure of early neolithic population in the South Caucasus
Communications Biology (2023)
-
Early contact between late farming and pastoralist societies in southeastern Europe
Nature (2023)
-
Process and Dynamics of Mediterranean Neolithization (7000–5500 bc)
Journal of Archaeological Research (2022)