Abstract
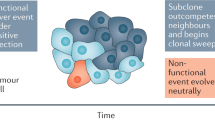
Intratumour heterogeneity and phenotypic plasticity, sustained by a range of somatic aberrations, as well as epigenetic and metabolic adaptations, are the principal mechanisms that enable cancers to resist treatment and survive under environmental stress. A comprehensive picture of the interplay between different somatic aberrations, from point mutations to whole-genome duplications, in tumour initiation and progression is lacking. We posit that different genomic aberrations generally exhibit a temporal order, shaped by a balance between the levels of mutations and selective pressures. Repeat instability emerges first, followed by larger aberrations, with compensatory effects leading to robust tumour fitness maintained throughout the tumour progression. A better understanding of the interplay between genetic aberrations, the microenvironment, and epigenetic and metabolic cellular states is essential for early detection and prevention of cancer as well as development of efficient therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
14 December 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41576-020-00313-9
References
American Cancer Society. Cancer facts & figures 2019. American Cancer Society www.cancer.org (2019).
Vasan, N., Baselga, J. & Hyman, D. M. A view on drug resistance in cancer. Nature 575, 299–309 (2019).
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Faltas, B. M. et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat. Genet. 48, 1490–1499 (2016).
Hu, Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019).
Nayar, U. et al. Acquired HER2 mutations in ER+ metastatic breast cancer confer resistance to estrogen receptor-directed therapies. Nat. Genet. 51, 207–216 (2019).
Ye, K. et al. Systematic discovery of complex insertions and deletions in human cancers. Nat. Med. 22, 97–104 (2016).
Wooster, R. et al. Instability of short tandem repeats (microsatellites) in human cancers. Nat. Genet. 6, 152–156 (1994).
Hause, R. J., Pritchard, C. C., Shendure, J. & Salipante, S. J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22, 1342–1350 (2016).
Campbell, B. B. et al. Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056.e10 (2017).
Persi, E. et al. Proteomic and genomic signatures of repeat instability in cancer and adjacent normal tissues. Proc. Natl Acad. Sci. USA 116, 16987–16996 (2019).
Winge, Ö. Zytologische untersuchungen über die natur maligner tumoren [German]. Z. Zellforsch. 10, 683–735 (1930).
Abdel-Rahman, W. M. et al. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl Acad. Sci. USA 98, 2538–2543 (2001).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203 (2012).
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 15, 139–150 (2018).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018).
Ben-David, U. & Amon, A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2020).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018).
Sniegowski, P. D., Gerrish, P. J. & Lenski, R. E. Evolution of high mutation rates in experimental populations of E. coli. Nature 387, 703–705 (1997).
Lang, G. I. et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Yizhak, K. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019).
Tomasetti, C. & Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015).
Tomasetti, C., Li, L. & Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017).
Reynolds, T. Y., Rockwell, S. & Glazer, P. M. Genetic instability induced by the tumor microenvironment. Cancer Res. 56, 5754–5757 (1996).
Koshiji, M. et al. HIF-1α induces genetic instability by transcriptionally downregulating MutSα expression. Mol. Cell 17, 793–803 (2005).
Gillies, R. J., Verduzco, D. & Gatenby, R. A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 12, 487–493 (2012).
Alexandrov, L. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Lipinski, K. A. et al. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer 2, 49–63 (2016).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Gillies, R. J., Brown, J. S., Anderson, A. R. A. & Gatenby, R. A. Eco-evolutionary causes and consequences of temporal changes in intratumoural blood flow. Nat. Rev. Cancer 18, 576–585 (2018).
DeBerardinis, R. J. & Chandel, N. S. Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016).
Feinberg, A. P., Koldobskiy, M. A. & Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 (2016).
Kinnaird, A., Zhao, S., Wellen, K. E. & Michelakis, E. D. Metabolic control of epigenetics in cancer. Nat. Rev. Cancer 16, 694–707 (2016).
Gatenby, R. A. & Gillies, R. J. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer 8, 56–61 (2008).
Gatenby, R. A., Gillies, R. J. & Brown, J. S. Of cancer and cave fish. Nat. Rev. Cancer 11, 237–238 (2011).
Lloyd, M. C. et al. Darwinian dynamics of intratumoral heterogeneity: not solely random mutations but also variable environmental selection forces. Cancer Res. 76, 3136–3144 (2016).
Maley, C. C. et al. Classifying the evolutionary and ecological features of neoplasms. Nat. Rev. Cancer 17, 605–619 (2017).
Lynch, M. & Conery, J. S. The origins of genome complexity. Science 302, 1401–1404 (2003).
Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, 1983).
Lynch, M. The Origins of Genome Architecture (Sinauer Associates (1723), 2007).
Koonin, E. V. The Logic of Chance: The Nature and Origin of Biological Evolution 1st edn (FT Press Science, 2012).
Muller, H. J. The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 (1964).
Haigh, J. The accumulation of deleterious genes in a population — Muller’s ratchet. Theor. Popul. Biol. 14, 251–267 (1978).
Dolja, V. V. & Koonin, E. V. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 1, 322–331 (2011).
Reuter, S. et al. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl Acad. Sci. USA 111, 6768–6773 (2014).
Kimura, M. On the probability of fixation of mutant genes in a population. Genetics 47, 713–719 (1962).
Kryazhimskiy, S. & Plotkin, J. B. The population genetics of dN/dS. PLoS Genet. 4, e1000304 (2008).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Futreal, P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Goyal, S. et al. Dynamic mutation-selection balance as an evolutionary attractor. Genetics 191, 1309–1319 (2012).
McFarland, C. D., Korolev, K. S., Kryukov, G. V., Sunyaev, S. R. & Mirny, L. A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl Acad. Sci. USA 110, 2910–2915 (2013).
McFarland, C. D., Mirny, L. A. & Korolev, K. S. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc. Natl Acad. Sci. USA 111, 15138–15143 (2014).
Cox, D. R. Regression models and life-tables. J. R. Stat. Soc. B 34, 187–220 (1972).
Persi, E., Wolf, Y. I., Leiserson, M. D. M., Koonin, E. V. & Ruppin, E. Criticality in tumor evolution and clinical outcome. Proc. Natl Acad. Sci. USA 115, E11101–E11110 (2018).
McFarland, C. D. et al. The damaging effect of passenger mutations on cancer progression. Cancer Res. 77, 4763–4772 (2017).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Weghorn, D. & Sunyaev, S. Bayesian inference of negative and positive selection in human cancers. Nat. Genet. 49, 1785–1788 (2017).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Williams, M. J. et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nat. Genet. 50, 895–903 (2018).
Kumar, S. et al. Passenger mutations in more than 2,500 cancer genomes: overall molecular functional impact and consequences. Cell 180, 915–927.e16 (2020).
López, S. et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat. Genet. 52, 283–293 (2020).
Kashi, Y. & King, D. G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22, 253–259 (2006).
Gemayel, R., Vinces, M. D., Legendre, M. & Verstrepen, K. J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Ann. Rev. Genet. 44, 445–477 (2010).
Chavali, S. et al. Constraints and consequences of the emergence of amino acid repeats in eukaryotic proteins. Nat. Struct. Mol. Biol. 24, 765–777 (2017).
Levinson, G. & Gutman, G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203–221 (1987).
Charlesworth, B., Sniegowski, P. & Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220 (1994).
Pâques, F., Leung, W. Y. & Haber, J. E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell Biol. 18, 2045–2054 (1998).
Hoelz, A., Debler, E. W. & Blobel, G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613–643 (2011).
Pick, E., Hofmann, K. & Glickman, M. H. PCI complexes: beyond the proteasome, CSN, and eIF3 troika. Mol. Cell 35, 260–264 (2009).
Klug, A. & Rhodes, D. ‘Zinc fingers’: a novel protein motif for nucleic acid recognition. Trends Biochem. Sci. 12, 464–469 (1987).
Neer, E. J., Schmidt, C. J., Nambudripad, R. & Smith, T. F. The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300 (1994).
Bell, J. K. et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24, 528–533 (2003).
Mosavi, L. K., Cammett, T. J., Desrosiers, D. C. & Peng, Z. Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 (2004).
Chavali, S., Singh, A. K., Santhanam, B. & Babu, M. Amino acid homorepeats in proteins. Nat. Rev. Chem. 4, 420–434 (2020).
Verstrepen, K. J., Jansen, A., Lewitter, F. & Fink, G. R. Intragenic tandem repeats generate functional variability. Nat. Genet. 37, 986–990 (2005).
Fondon, J. W. 3rd & Garner, H. R. Molecular origins of rapid and continuous morphological evolution. Proc. Natl Acad. Sci. USA 101, 18058–18063 (2004).
Sawyer, L. A. et al. Natural variation in a Drosophila clock gene and temperature compensation. Science 278, 2117–2120 (1997).
Gymrek, M. et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 48, 22–29 (2016).
Santos-Pereira, J. M. & Aguilera, A. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 (2015).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Bibel, M. & Barde, Y. A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 14, 2919–2937 (2000).
Nithianantharajah, J. & Hannan, A. J. Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. Bioessays 29, 525–535 (2007).
Kannan, M. et al. WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc. Natl Acad. Sci. USA 114, E9308–E9317 (2017).
Hammock, E. A. & Young, L. J. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308, 1630–1634 (2005).
Segurel, L., Leffler, E. M. & Przeworski, M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 9, e1001211 (2011).
Davies, B. et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530, 171–176 (2016).
Persi, E., Wolf, Y. I. & Koonin, E. V. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat. Commun. 7, 13570 (2016).
Schaper, E., Gascuel, O. & Anisimova, M. Deep conservation of human protein tandem repeats within the eukaryotes. Mol. Biol. Evol. 31, 1132–1148 (2014).
Marcotte, E. M., Pellegrini, M., Yeates, T. O. & Eisenberg, D. A census of protein repeats. J. Mol. Biol. 293, 151–160 (1999).
Persi, E. & Horn, D. Systematic analysis of compositional order of proteins reveals new characteristics of biological functions and a universal correlate of macroevolution. PLoS Comput. Biol. 9, e1003346 (2013).
Karlin, S., Brocchieri, L., Bergman, A., Mrazek, J. & Gentles, A. J. Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl Acad. Sci. USA 99, 333–338 (2002).
Gatchel, J. R. & Zoghbi, H. Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6, 743–755 (2005).
Pearson, C. E., Nichol Edamura, K. & Cleary, J. D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 (2005).
Mirkin, S. M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
La Spada, A. R. & Taylor, J. P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11, 247–258 (2010).
López Castel, A., Cleary, J. D. & Pearson, C. E. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11, 165–170 (2010).
Carvalho, C. M. & Lupski, J. R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224–238 (2016).
Hannan, A. J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 19, 286–298 (2018).
Khristich, A. N. & Mirkin, S. M. On the wrong DNA track: molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 295, 4134–4170 (2020).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Ishiura, H. et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 50, 581–590 (2018).
Loeb, L. A., Springgate, C. F. & Battula, N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 34, 2311–2321 (1974).
Duval, A. & Hamelin, R. Mutations at coding repeat sequences in mismatch repair deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 62, 2447–2454 (2002).
Treangen, T. J. & Salzberg, S. L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46 (2011).
El-Metwally, S., Hamza, T., Zakaria, M. & Helmy, M. Next-generation sequence assembly: four stages of data processing and computational challenges. PLoS Comput. Biol. 9, e1003345 (2013).
Popat, S., Hubner, R. & Houlston, R. S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 23, 609–618 (2005).
Li, B. et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 48, 725–732 (2016).
Germano, G. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Yarchoan, M., Johnson, B. A. 3rd, Lutz, E. R., Laheru, D. A. & Jaffee, E. M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 17, 209–222 (2017).
Mlecnik, B. et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44, 698–711 (2016).
Mlecnik, B. et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci. Transl. Med. 8, 327ra26 (2016).
Ionov, Y., Peinado, M. A., Malkhosyan, S., Shibata, D. & Perucho, M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363, 558–561 (1993).
Torres, E. M. et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924 (2007).
Pavelka, N. et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325 (2010).
Weaver, B. A., Silk, A. D., Montagna, C., Verdier-Pinard, P. & Cleveland, D. W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007).
Rowald, K. et al. Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep. 15, 2679–2691 (2016).
Sotillo, R., Schvartzman, J. M., Socci, N. D. & Benezra, R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464, 436–440 (2010).
Bakhoum, S. F., Ngo, B., Laughney, A. M., Cavallo, J. A. & Murphy, C. J. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018).
Gao, C. et al. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl Acad. Sci. USA 113, 14793–14798 (2016).
Danielsen, H. E., Pradhan, M. & Novelli, M. Revisiting tumour aneuploidy — the place of ploidy assessment in the molecular era. Nat. Rev. Clin. Oncol. 13, 291–304 (2016).
Auslander, N., Wolf, Y. I. & Koonin, E. V. Interplay between DNA damage repair and apoptosis shapes cancer evolution through aneuploidy and microsatellite instability. Nat. Commun. 11, 1234 (2020).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Roylance, R. et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomarkers Prev. 20, 2183–2194 (2011).
Zasadil, L. M. et al. High rates of chromosome missegregation suppress tumor progression but do not inhibit tumor initiation. Mol. Biol. Cell 27, 1981–1989 (2016).
Silk, A. D. et al. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl Acad. Sci. USA 110, E4134–E4141 (2013).
Sansregret, L. et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 7, 218–233 (2017).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Díaz-Carballo, D. et al. A distinct oncogenerative multinucleated cancer cell serves as a source of stemness and tumor heterogeneity. Cancer Res. 78, 2318–2331 (2018).
Pienta, K. J., Hammarlund, E. U., Axelrod, R., Brown, J. S. & Amend, S. R. Poly-aneuploid cancer cells promote evolvability, generating lethal cancer. Evolut. Appl. 13, 1626–1634 (2020).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Smith, J. C. & Sheltzer, J. M. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 7, e39217 (2018).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000).
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I. & Koonin, E. V. Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008 (2002).
Innan, H. & Kondrashov, F. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108 (2010).
Dewhurst, S. M. et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 4, 175–185 (2014).
Maciver, S. K. Asexual amoebae escape Muller’s ratchet through polyploidy. Trends Parasitol. 32, 855–862 (2016).
Ford, A. M. et al. Fetal origins of the TEL–AML1 fusion gene in identical twins with leukemia. Proc. Natl Acad. Sci. USA 95, 4584–4588 (1998).
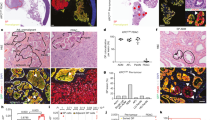
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020).
Rausch, T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012).
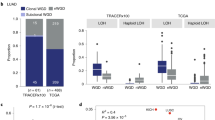
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 173, 611–623.e17 (2018).
Cortés-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 52, 331–341 (2020).
Paulson, T. G. et al. Chromosomal instability and copy number alterations in Barrett’s esophagus and esophageal adenocarcinoma. Clin. Cancer Res. 15, 3305–3314 (2009).
Martinez, P. et al. Evolution of Barrett’s esophagus through space and time at single-crypt and whole-biopsy levels. Nat. Commun. 9, 794 (2018).
Ordway, B., Swietach, P., Gillies, R. J. & Damaghi, M. Causes and consequences of variable tumor cell metabolism on heritable modifications and tumor evolution. Front. Oncol. 10, 373 (2020).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Gillies, R. J., Robey, I. & Gatenby, R. A. Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 49 (Suppl. 2), 24S–42S (2008).
Webb, B. A., Chimenti, M., Jacobson, M. P. & Barber, D. L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011).
Corbet, C. & Feron, O. Tumour acidosis: from the passenger to the driver’s seat. Nat. Rev. Cancer 17, 577–593 (2017).
Brand, A. et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 24, 657–671 (2016).
Pilon-Thomas, S. et al. Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Res. 76, 1381–1390 (2016).
Persi, E. et al. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat. Commun. 9, 2997 (2018).
Hoffmann, A. A. & Hercus, M. J. Environmental stress as an evolutionary force. BioScience 50, 217–226 (2000).
Ram, Y. & Hadany, L. The evolution of stress-induced hypermutation in asexual populations. Evolution 66, 2315–2328 (2012).
McKenzie, G. J., Harris, R. S., Lee, P. L. & Rosenberg, S. M. The SOS response regulates adaptive mutation. Proc. Natl Acad. Sci. USA 97, 6646–6651 (2000).
Rattray, A. J. & Strathern, J. N. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 37, 31–66 (2003).
Krutyakov, V. M. Eukaryotic error-prone DNA polymerases: the presumed roles in replication, repair, and mutagenesis. Mol. Biol. 40, 1–8 (2006).
Fitzgerald, D. M., Hastings, P. J. & Rosenberg, S. M. Stress-induced mutagenesis: implications in cancer and drug resistance. Annu. Rev. Cancer Biol. 1, 119–140 (2017).
Clara, J. A., Monge, C., Yang, Y. & Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells — a clinical update. Nat. Rev. Clin. Oncol. 17, 204–232 (2020).
Yun, J. et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 325, 1555–1559 (2009).
Young, S. D., Marshall, R. S. & Hill, R. P. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl Acad. Sci. USA 85, 9533–9537 (1988).
Jackson, A. L., Chen, R. & Loeb, A. L. Induction of microsatellite instability by oxidative DNA damage. Proc. Natl Acad. Sci. USA 95, 12468–12473 (1998).
Chatterjee, N., Lin, Y., Santillan, B. A., Yotnda, P. & Wilson, J. H. Environmental stress induces trinucleotide repeat mutagenesis in human cells. Proc. Natl Acad. Sci. USA 112, 3764–3769 (2015).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013).
Matzke, M. A., Mittelsten Scheid, O. & Matzke, A. J. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays 21, 761–767 (1999).
Tang, Y. C. & Amon, A. Gene copy-number alterations: a cost–benefit analysis. Cell 152, 394–405 (2013).
Herrera, L. A., Prada, D., Andonegui, M. A. & Dueñas-González, A. The epigenetic origin of aneuploidy. Curr. Genomics 9, 43–50 (2008).
Chen, G., Bradford, W. D., Seidel, C. W. & Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250 (2012).
Yona, A. H. et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl Acad. Sci. USA 109, 21010–21015 (2012).
Espina, V. & Liotta, L. A. What is the malignant nature of human ductal carcinoma in situ? Nat. Rev. Cancer 11, 68–75 (2011).
Filippova, G. N. et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28, 335–343 (2001).
Libby, R. T. et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 4, e1000257 (2008).
Dion, V., Lin, Y., Hubert, L. Jr, Waterland, R. A. & Wilson, J. H. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum. Mol. Genet. 17, 1306–1317 (2008).
Gallagher, E. J. & LeRoith, D. Obesity and diabetes: the increased risk of cancer and cancer-related mortality. Physiol. Rev. 95, 727–748 (2015).
Kussell, E. & Leibler, S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078 (2005).
Cvijović, I., Good, B. H., Jerison, E. R. & Desai, M. M. Fate of a mutation in a fluctuating environment. Proc. Natl Acad. Sci. USA 112, E5021–E5028 (2015).
Gatenby, R. A., Cunningham, J. J. & Brown, J. S. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat. Commun. 5, 5499 (2014).
Schiffman, J. D., White, R. M., Graham, T. A., Huang, Q. & Aktipis, A. in Frontiers in Cancer Research: Evolutionary Foundations, Revolutionary Directions (eds. C. C. Maley & M. Greaves) 135–176 (Springer, 2016).
Ashworth, A., Lord, C. J. & Reis-Filho, J. S. Genetic interactions in cancer progression and treatment. Cell 145, 30–38 (2011).
Phillips, P. C. Epistasis — the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 (2008).
Poelwijk, F. J., Tănase-Nicola, S., Kiviet, D. J. & Tans, S. J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 272, 141–144 (2011).
O’Neil, N. J., Bailey, M. L. & Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 18, 613–623 (2017).
Huang, A., Garraway, L. A., Ashworth, A. & Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 19, 23–38 (2020).
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Bitler, B. G. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 21, 231–238 (2015).
Chan, E. M. et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019).
Trachootham, D., Alexandre, J. & Huang, P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Dis. 8, 579–591 (2009).
Acknowledgements
The authors thank members of the Koonin group at the National Center for Biotechnology Information (NCBI) and members of the Moffitt Cancer Center for useful discussions and feedback. E.P. and E.V.K. are supported by intramural funds from the US Department of Health and Human Services (to the National Library of Medicine). R.J.G. and R.A.G. acknowledge support from the Physical Sciences in Oncology Program U54CA193489. F.D. is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 648670).
Author information
Authors and Affiliations
Contributions
E.P. researched data for the article. E.P. and E.V.K. wrote the article and reviewed/edited the manuscript before submission. All authors substantially contributed to discussion of content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks P. Campbell, C. Maley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Persi, E., Wolf, Y.I., Horn, D. et al. Mutation–selection balance and compensatory mechanisms in tumour evolution. Nat Rev Genet 22, 251–262 (2021). https://doi.org/10.1038/s41576-020-00299-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-020-00299-4
This article is cited by
-
Machine learning-based extrachromosomal DNA identification in large-scale cohorts reveals its clinical implications in cancer
Nature Communications (2024)
-
Protein degradation: expanding the toolbox to restrain cancer drug resistance
Journal of Hematology & Oncology (2023)
-
Deep mutational scanning reveals the molecular determinants of RNA polymerase-mediated adaptation and tradeoffs
Nature Communications (2023)
-
Tumour immunotherapy: lessons from predator–prey theory
Nature Reviews Immunology (2022)
-
Targeting IL8 as a sequential therapy strategy to overcome chemotherapy resistance in advanced gastric cancer
Cell Death Discovery (2022)