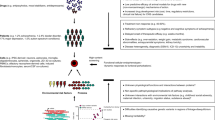
Abstract
In spite of major efforts and investment in development of psychiatric drugs, many clinical trials have failed in recent decades, and clinicians still prescribe drugs that were discovered many years ago. Although multiple reasons have been discussed for the drug development deadlock, we focus here on one of the major possible biological reasons: differences between the characteristics of drug targets in preclinical models and the corresponding targets in patients. Importantly, based on technological advances in single-cell analysis, we propose here a framework for the use of available and newly emerging knowledge from single-cell and spatial omics studies to evaluate and potentially improve the translational predictivity of preclinical models before commencing preclinical and, in particular, clinical studies. We believe that these recommendations will improve preclinical models and the ability to assess drugs in clinical trials, reducing failure rates in expensive late-stage trials and ultimately benefitting psychiatric drug discovery and development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Howes, O. D. & Baxter, L. The drug treatment deadlock in psychiatry and the route forward. World Psychiatry 22, 2–3 (2023).
Marder, S. R., Laughren, T. & Romano, S. J. Why are innovative drugs failing in phase III? Am. J. Psychiatry 174, 829–831 (2017).
Spark, D. L., Fornito, A., Langmead, C. J. & Stewart, G. D. Beyond antipsychotics: a twenty-first century update for preclinical development of schizophrenia therapeutics. Transl. Psychiatry 12, 147 (2022).
Tricklebank, M. D., Robbins, T. W., Simmons, C. & Wong, E. H. F. Time to re-engage psychiatric drug discovery by strengthening confidence in preclinical psychopharmacology. Psychopharmacology 238, 1417–1436 (2021).
Correll, C. U. et al. The future of psychopharmacology: a critical appraisal of ongoing phase 2/3 trials, and of some current trends aiming to de-risk trial programmes of novel agents. World Psychiatry 22, 48–74 (2023).
Krystal, A. D. et al. The first implementation of the NIMH FAST-FAIL approach to psychiatric drug development. Nat. Rev. Drug. Discov. 18, 82–84 (2018).
Sarter, M. & Tricklebank, M. Revitalizing psychiatric drug discovery. Nat. Rev. Drug. Discov. 11, 423–424 (2012).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021).
Gouwens, N. W. et al. Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell 183, 935–953.e19 (2020).
Scala, F. et al. Phenotypic variation of transcriptomic cell types in mouse motor cortex. Nature 598, 144–150 (2021).
Clark, I. C. et al. Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 372, eabf1230 (2021).
Albert, P. R. & Le François, B. Modifying 5-HT1A receptor gene expression as a new target for antidepressant therapy. Front. Neurosci. 4, 35 (2010).
Negi, S. K. & Guda, C. Global gene expression profiling of healthy human brain and its application in studying neurological disorders. Sci. Rep. 7, 897 (2017).
Sjöstedt, E. et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 (2020).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Sinkeviciute, I. et al. Efficacy of different types of cognitive enhancers for patients with schizophrenia: a meta-analysis. NPJ Schizophr. 4, 22 (2018).
Schmidt, E. R. E. et al. A human-specific modifier of cortical connectivity and circuit function. Nature 599, 640–644 (2021).
Defelipe, J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front. Neuroanat. 5, 29 (2011).
Sousa, A. M. M., Meyer, K. A., Santpere, G., Gulden, F. O. & Sestan, N. Evolution of the human nervous system function, structure, and development. Cell 170, 226–247 (2017).
Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018).
Franjic, D. et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron 110, 452–469.e14 (2022).
Jorstad, N. L. et al. Transcriptomic cytoarchitecture reveals principles of human neocortex organization. Science 382, eadf6812 (2023).
Agarwal, D. et al. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat. Commun. 11, 4182 (2020).
Tran, M. N. et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron 109, 3088–3103.e5 (2021).
Sorrells, S. F. et al. Immature excitatory neurons develop during adolescence in the human amygdala. Nat. Commun. 10, 2748 (2019).
Masuda, T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Winkler, E. A. et al. A single-cell atlas of the normal and malformed human brain vasculature. Science 375, eabi7377 (2022).
Batiuk, M. Y. et al. Upper cortical layer-driven network impairment in schizophrenia. Sci. Adv. 8, eabn8367 (2022).
Haney, J. R. et al. Broad transcriptomic dysregulation across the cerebral cortex in ASD. Nature 611, 532–539 (2022).
Nagy, C. et al. Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat. Neurosci. 23, 771–781 (2020).
Howes, O. D., Thase, M. E. & Pillinger, T. Treatment resistance in psychiatry: state of the art and new directions. Mol. Psychiatry 27, 58–72 (2021).
Planert, H. et al. Cellular and synaptic diversity of layer 2-3 pyramidal neurons in human individuals. Preprint at https://doi.org/10.1101/2021.11.08.467668 (2023).
Johansen, N. et al. Interindividual variation in human cortical cell type abundance and expression. Science 382, eadf2359 (2023).
Asp, M., Bergenstråhle, J. & Lundeberg, J. Spatially resolved transcriptomes—next generation tools for tissue exploration. BioEssays 42, 1900221 (2020).
Fang, R. et al. Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science 377, 56–62 (2022).
Pfisterer, U. et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 11, 5038 (2020).
Petukhov, V. et al. Case-control analysis of single-cell RNA-seq studies. Preprint at https://doi.org/10.1101/2022.03.15.484475 (2022).
Fischer, D. S., Schaar, A. C. & Theis, F. J. Modeling intercellular communication in tissues using spatial graphs of cells. Nat. Biotechnol. 41, 332–336 (2023).
Campagnola, L. et al. Local connectivity and synaptic dynamics in mouse and human neocortex. Science 375, eabj5861 (2022).
Berg, J. et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021).
Kalmbach, B. E. et al. Signature morpho-electric, transcriptomic, and dendritic properties of human layer 5 neocortical pyramidal neurons. Neuron 109, 2914–2927.e5 (2021).
Lee, B. R. et al. Signature morphoelectric properties of diverse GABAergic interneurons in the human neocortex. Science 382, eadf6484 (2023).
Artigas, F. et al. Defining the brain circuits involved in psychiatric disorders: IMI-NEWMEDS. Nat. Rev. Drug. Discov. 16, 1–2 (2016).
Krystal, A. D. et al. A randomized proof-of-mechanism trial applying the ‘fast-fail’ approach to evaluating κ-opioid antagonism as a treatment for anhedonia. Nat. Med. 26, 760–768 (2020).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Perkel, J. M. Single-cell analysis enters the multiomics age. Nature 595, 614–616 (2021).
Kharchenko, P. V. The triumphs and limitations of computational methods for scRNA-seq. Nat. Methods 18, 723–732 (2021).
No authors listed.Method of the Year 2019: single-cell multimodal omics. Nat. Methods 17, 1 (2020).
Yuste, R. et al. A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat. Neurosci. 23, 1456–1468 (2020).
Bhaduri, A. et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021).
Callaway, E. M. et al. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Ziffra, R. S. et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021).
Zhong, S. et al. Decoding the development of the human hippocampus. Nature 577, 531–536 (2020).
Regev, A. et al. The human cell atlas. eLife 6, e27041 (2017).
Herring, C. A. et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447.e28 (2022).
Wheeler, M. A. et al. MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020).
Yang, A. C. et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571 (2021).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497–514.e22 (2020).
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991.e19 (2020).
Li, Y. E. et al. An atlas of gene regulatory elements in adult mouse cerebrum. Nature 598, 129–136 (2021).
Kozareva, V. et al. A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature 598, 214–219 (2021).
Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021).
Di Bella, D. J. et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature 595, 554–559 (2021).
La Manno, G. et al. Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021).
Romanov, R. A. et al. Molecular design of hypothalamus development. Nature 582, 246–252 (2020).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Lui, J. H. et al. Differential encoding in prefrontal cortex projection neuron classes across cognitive tasks. Cell 184, 489–506.e26 (2021).
Malwade, S. et al. Identification of vulnerable interneuron subtypes in 15q13.3 microdeletion syndrome using single-cell transcriptomics. Biol. Psychiatry 91, 727–739 (2022).
Gouwens, N. W. et al. Classification of electrophysiological and morphological neuron types in the mouse visual cortex. Nat. Neurosci. 22, 1182–1195 (2019).
Joglekar, A. et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12, 463 (2021).
Bandler, R. C. et al. Single-cell delineation of lineage and genetic identity in the mouse brain. Nature 601, 404–409 (2022).
Peng, H. et al. Morphological diversity of single neurons in molecularly defined cell types. Nature 598, 174–181 (2021).
Liu, H. et al. DNA methylation atlas of the mouse brain at single-cell resolution. Nature 598, 120–128 (2021).
Yao, Z. et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110 (2021).
Krienen, F. M. et al. Innovations present in the primate interneuron repertoire. Nature 586, 262–269 (2020).
Bakken, T. E. et al. Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans. eLife 10, e64875 (2021).
Borm, L. E. et al. Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH. Nat. Biotechnol. 41, 222–231 (2022).
Yu, Y. et al. Interneuron origin and molecular diversity in the human fetal brain. Nat. Neurosci. 24, 1745–1756 (2021).
Shi, Y. et al. Mouse and human share conserved transcriptional programs for interneuron development. Science 374, eabj6641 (2021).
Eze, U. C., Bhaduri, A., Haeussler, M., Nowakowski, T. J. & Kriegstein, A. R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 24, 584–594 (2021).
Close, J. L. et al. Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron 93, 1035–1048 e5 (2017).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016).
Samudyata et al. SARS-CoV-2 promotes microglial synapse elimination in human brain organoids. Mol. Psychiatry 27, 3939–3950 (2022).
Yang, S. M., Michel, K., Jokhi, V., Nedivi, E. & Arlotta, P. Neuron class-specific responses govern adaptive myelin remodeling in the neocortex. Science 370, eabd2109 (2020).
Hao, Z.-Z. et al. Single-cell transcriptomics of adult macaque hippocampus reveals neural precursor cell populations. Nat. Neurosci. 25, 805–817 (2022).
Lei, Y. et al. Spatially resolved gene regulatory and disease-related vulnerability map of the adult macaque cortex. Nat. Commun. 13, 1–20 (2022).
Khodosevich, K. & Sellgren, C. M. Neurodevelopmental disorders—high-resolution rethinking of disease modeling. Mol. Psychiatry 28, 34–43 (2023).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Derks, J. et al. Increasing the throughput of sensitive proteomics by plexDIA. Nat. Biotechnol. 41, 50–59 (2023).
Huffman, R. G. et al. Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics. Nat. Methods 20, 714–722 (2023).
Specht, H. et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 22, 50 (2021).
Mohan, H. et al. Dendritic and axonal architecture of individual pyramidal neurons across layers of adult human neocortex. Cereb. Cortex 25, 4839–4853 (2015).
Loomba, S. et al. Connectomic comparison of mouse and human cortex. Science 377, eabo0924 (2022).
Zhu, F., Nair, R. R., Fisher, E. M. C. & Cunningham, T. J. Humanising the mouse genome piece by piece. Nat. Commun. 10, 1845 (2019).
Normand, R. et al. Found in translation: a machine learning model for mouse-to-human inference. Nat. Methods 15, 1067–1073 (2018).
Drysdale, A. T. et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med. 23, 28–38 (2017).
Contractor, A., Klyachko, V. A. & Portera-Cailliau, C. Altered neuronal and circuit excitability in fragile X syndrome. Neuron 87, 699–715 (2015).
Oyama, K. & Sakatani, K. Machine learning-based assessment of cognitive impairment using time-resolved near-infrared spectroscopy and basic blood test. Front. Neurol. 12, 2641 (2022).
Wu, Y. E., Pan, L., Zuo, Y., Li, X. & Hong, W. Detecting activated cell populations using single-cell RNA-seq. Neuron 96, 313–329.e6 (2017).
Lacar, B. et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022 (2016).
Clancy, B., Darlington, R. B. & Finlay, B. L. Translating developmental time across mammalian species. Neuroscience 105, 7–17 (2001).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Wu, J. et al. Integrating spatial and single-nucleus transcriptomic data elucidates microglial-specific responses in female cynomolgus macaques with depressive-like behaviors. Nat. Neurosci. 26, 1352–1364 (2023).
Piñero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Zhang, M. J. et al. Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 54, 1572–1580 (2022).
Vasistha, N. A. & Khodosevich, K. The impact of (ab)normal maternal environment on cortical development. Prog. Neurobiol. 202, 102054 (2021).
Wang, Y. Y. et al. CeDR Atlas: a knowledgebase of cellular drug response. Nucleic Acids Res. 50, D1164–D1171 (2022).
Schoof, E. M. et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 (2021).
Karlsson, M. et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169 (2021).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Ball, G. et al. Cortical morphology at birth reflects spatiotemporal patterns of gene expression in the fetal human brain. PLoS Biol. 18, e3000976 (2020).
Dachet, F. et al. Selective time-dependent changes in activity and cell-specific gene expression in human postmortem brain. Sci. Rep. 11, 6078 (2021).
Kaar, S. J., Natesan, S., McCutcheon, R. & Howes, O. D. Antipsychotics: mechanisms underlying clinical response and side-effects and novel treatment approaches based on pathophysiology. Neuropharmacology 172, 107704 (2020).
Lobo, M. C., Whitehurst, T. S., Kaar, S. J. & Howes, O. D. New and emerging treatments for schizophrenia: a narrative review of their pharmacology, efficacy and side effect profile relative to established antipsychotics. Neurosci. Biobehav. Rev. 132, 324–361 (2022).
Mizuno, Y., McCutcheon, R. A., Brugger, S. P. & Howes, O. D. Heterogeneity and efficacy of antipsychotic treatment for schizophrenia with or without treatment resistance: a meta-analysis. Neuropsychopharmacology 45, 622–631 (2019).
Lee, B. et al. Signature morphoelectric properties of diverse GABAergic interneurons in the human neocortex. Science 382, eadf6484 (2023).
Molnár, G. et al. Complex events initiated by individual spikes in the human cerebral cortex. PLOS Biol. 6, e222 (2008).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Bakken, T. E. et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS ONE 13, e0209648 (2018).
Cadwell, C. R. et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Changeux, J. P. The nicotinic acetylcholine receptor: the founding father of the pentameric ligand-gated ion channel superfamily. J. Biol. Chem. 287, 40207–40215 (2012).
Broide, R. S., Winzer-Serhan, U. H., Chen, Y. & Leslie, F. M. Distribution of α7 nicotinic acetylcholine receptor subunit mRNA in the developing mouse. Front. Neuroanat. 13, 76 (2019).
Wu, J. et al. Heteromeric α7β2 nicotinic acetylcholine receptors in the brain. Trends Pharmacol. Sci. 37, 562–574 (2016).
Dineley, K. T., Pandya, A. A. & Yakel, J. L. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol. Sci. 36, 96–108 (2015).
Leonard, S. et al. Association of promoter variants in the α7 nicotinic acetylcholine receptor subunit gene with an inhibitory deficit found in schizophrenia. Arch. Gen. Psychiatry 59, 1085–1096 (2002).
Stephens, S. H. et al. Association of the 5′-upstream regulatory region of the α7 nicotinic acetylcholine receptor subunit gene (CHRNA7) with schizophrenia. Schizophr. Res. 109, 102–112 (2009).
Terry, A. V. & Callahan, P. M. α7 Nicotinic acetylcholine receptors as therapeutic targets in schizophrenia: update on animal and clinical studies and strategies for the future. Neuropharmacology 170, 108053 (2020).
Tregellas, J. R. & Wylie, K. P. Alpha7 nicotinic receptors as therapeutic targets in schizophrenia. Nicotine Tob. Res. 21, 349–356 (2019).
Nichols, D. E. & Nichols, C. D. Serotonin receptors. Chem. Rev. 108, 1614–1641 (2008).
Berger, M., Gray, J. A. & Roth, B. L. The expanded biology of serotonin. Annu. Rev. Med. 60, 355–366 (2009).
Meltzer, H. Y. & Massey, B. W. The role of serotonin receptors in the action of atypical antipsychotic drugs. Curr. Opin. Pharmacol. 11, 59–67 (2011).
Zhang, G. & Stackman, R. W. The role of serotonin 5-HT2A receptors in memory and cognition. Front. Pharmacol. 6, 225 (2015).
Miyamoto, S., Miyake, N., Jarskog, L. F., Fleischhacker, W. W. & Lieberman, J. A. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol. Psychiatry 17, 1206–1227 (2012).
Ebdrup, B. H., Rasmussen, H., Arnt, J. & Glenthøj, B. Serotonin 2A receptor antagonists for treatment of schizophrenia. Expert Opin. Investig. Drugs 20, 1211–1223 (2011).
Pinard, E., Borroni, E., Koerner, A., Umbricht, D. & Alberati, D. Glycine transporter type I (GlyT1) inhibitor, bitopertin: a journey from lab to patient. Chimia 72, 477 (2018).
Bugarski-Kirola, D. et al. Bitopertin in negative symptoms of schizophrenia—results from the phase III flashlyte and daylyte studies. Biol. Psychiatry 82, 8–16 (2017).
Acknowledgements
Work in the Khodosevich lab is supported by Novo Nordisk Foundation Hallas-Møller Investigator grants (NNF16OC0019920 and NNF21OC0067146) and Lundbeck Foundation Ascending Investigator grant (2020-1025). Work in the Howes lab is funded by Medical Research Council UK (no. MC_A656_5QD30_2135), Maudsley Charity (no. 666) and Wellcome Trust (no. 094849/Z/10/Z). The authors thank S. Hopkins (Sunovion Pharmaceuticals) for comments on the initial version of the manuscript. We are grateful to O. Kharchenko for their work on figure illustrations.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
K.K. and K.D. have no potential conflicts of interest. O.H. is a part-time employee of Lundbeck A/s and has received investigator-initiated research funding from and/or participated in advisory or speaker meetings organized by Angellini, Autifony, Biogen, Boehringer-Ingelheim, Eli Lilly, Heptares, Global Medical Education, Invicro, Jansen, Lundbeck, Neurocrine, Otsuka, Sunovion, Rand, Recordati, Roche, Rovi and Viatris/Mylan. O.H. has a patent for the use of dopaminergic imaging.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks Wayne Drevets, Brisa Fernandes and Christopher Langmead for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Diffusion tensor imaging
-
(DTI). A structural MRI technique that estimates axonal tract organization based on anisotropic diffusion.
- Functional MRI
-
(fMRI). An imaging technique that measures brain activity using changes in blood flow.
- Induced pluripotent stem cell
-
(iPSC). Pluripotent cell that was reprogrammed from a somatic cell.
- Modality
-
In single-cell research, refers to the type of molecule that is being analysed, for example, RNA, DNA, methylation, open chromatin, protein and so forth.
- Positron emission tomography
-
(PET). An imaging technique that uses radioactive tracers to visualize and estimate distribution of specific molecules in the brain; in relation to psychiatric disorders, it can detect distinct receptor distribution, distribution of metabolic molecules and other parameters.
- Uniform manifold approximation and projection
-
(UMAP). A technique for dimensionality reduction to visualize single-cell omics data, where dimensionality is reduced from multiple to 2D.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khodosevich, K., Dragicevic, K. & Howes, O. Drug targeting in psychiatric disorders — how to overcome the loss in translation?. Nat Rev Drug Discov 23, 218–231 (2024). https://doi.org/10.1038/s41573-023-00847-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-023-00847-7