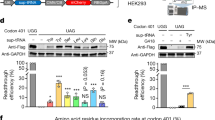
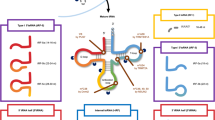
Abstract
Transfer RNAs (tRNAs) have a crucial role in protein synthesis, and in recent years, their therapeutic potential for the treatment of genetic diseases — primarily those associated with a mutation altering mRNA translation — has gained significant attention. Engineering tRNAs to readthrough nonsense mutation-associated premature termination of mRNA translation can restore protein synthesis and function. In addition, supplementation of natural tRNAs can counteract effects of missense mutations in proteins crucial for tRNA biogenesis and function in translation. This Review will present advances in the development of tRNA therapeutics with high activity and safety in vivo and discuss different formulation approaches for single or chronic treatment modalities. The field of tRNA therapeutics is still in its early stages, and a series of challenges related to tRNA efficacy and stability in vivo, delivery systems with tissue-specific tropism, and safe and efficient manufacturing need to be addressed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Huang, X. et al. The landscape of mRNA nanomedicine. Nat. Med. 28, 2273–2287 (2022).
Rohner, E., Yang, R., Foo, K. S., Goedel, A. & Chien, K. R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 40, 1586–1600 (2022).
Brown, A., Shao, S., Murray, J., Hegde, R. S. & Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015).
Crick, F. H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966).
Dittmar, K. A., Goodenbour, J. M. & Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006).
Kirchner, S. & Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112 (2015).
Polte, C. et al. Assessing cell-specific effects of genetic variations using tRNA microarrays. BMC Genom. 20, 549 (2019).
Thornlow, B. P. et al. Predicting transfer RNA gene activity from sequence and genome context. Genome Res. 30, 85–94 (2020). This study shows that a broader sequence from the tRNA loci affects tRNA expression that could instruct the selection of tissue-specific tRNA promoters.
Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).
Rak, R., Dahan, O. & Pilpel, Y. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu. Rev. Cell Dev. Biol. 34, 239–264 (2018).
Torrent, M., Chalancon, G., de Groot, N. S., Wuster, A. & Madan Babu, M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 11, eeat6409 (2018).
Mort, M., Ivanov, D., Cooper, D. N. & Chuzhanova, N. A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 29, 1037–1047 (2008).
Floquet, C., Hatin, I., Rousset, J. P. & Bidou, L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 8, e1002608 (2012).
Keeling, K. M., Xue, X., Gunn, G. & Bedwell, D. M. Therapeutics based on stop codon readthrough. Annu. Rev. Genom. Hum. Genet. 15, 371–394 (2014).
Spelier, S., van Doorn, E. P. M., van der Ent, C. K., Beekman, J. M. & Koppens, M. A. J. Readthrough compounds for nonsense mutations: bridging the translational gap. Trends Mol. Med. 29, 297–314 (2023).
Ryan, N. J. Ataluren: first global approval. Drugs 74, 1709–1714 (2014).
Capecchi, M. R. & Gussin, G. N. Suppression in vitro: identification of a serine-sRNA as a “nonsense” suppressor. Science 149, 417–422 (1965).
Hirsh, D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 58, 439–458 (1971).
Temple, G. F., Dozy, A. M., Roy, K. L. & Kan, Y. W. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for β-thalassaemia. Nature 296, 537–540 (1982).
Davyt, M., Bharti, N. & Ignatova, Z. Effect of mRNA/tRNA mutations on translation speed: implications for human diseases. J. Biol. Chem. 299, 105089 (2023).
Khorkova, O., Stahl, J., Joji, A., Volmar, C. H. & Wahlestedt, C. Amplifying gene expression with RNA-targeted therapeutics. Nat. Rev. Drug Discov. 22, 539–561 (2023).
Zhu, Y., Zhu, L., Wang, X. & Jin, H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
Anastassiadis, T. & Kohrer, C. Ushering in the era of tRNA medicines. J. Biol. Chem. 299, 105246 (2023).
Dolgin, E. tRNA therapeutics burst onto startup scene. Nat. Biotechnol. 40, 283–286 (2022).
Capecchi, M. R., Haar, R. A., Capecchi, N. E. & Sveda, M. M. The isolation of a suppressible nonsense mutant in mammalian cells. Cell 12, 371–381 (1977).
Chang, J. C., Temple, G. F., Trecartin, R. F. & Kan, Y. W. Suppression of the nonsense mutation in homozygous beta 0 thalassaemia. Nature 281, 602–603 (1979).
Bordeira-Carrico, R. et al. Rescue of wild-type E-cadherin expression from nonsense-mutated cancer cells by a suppressor-tRNA. Eur. J. Hum. Genet. 22, 1085–1092 (2014).
Buvoli, M., Buvoli, A. & Leinwand, L. A. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol. Cell Biol. 20, 3116–3124 (2000).
Kiselev, A. V. et al. [Suppression of nonsense mutations in the dystrophin gene by a suppressor tRNA gene]. Mol. Biol. 36, 43–47 (2002).
Laski, F. A., Belagaje, R., RajBhandary, U. L. & Sharp, P. A. An amber suppressor tRNA gene derived by site-specific mutagenesis: cloning and function in mammalian cells. Proc. Natl Acad. Sci. USA 79, 5813–5817 (1982).
Lueck, J. D. et al. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 10, 822 (2019). The study determines that the systematic alterations of the anticodon of different human tRNA isoacceptor families results in sup-tRNAs with diferent efficiency.
Panchal, R. G., Wang, S., McDermott, J. & Link, C. J. Jr. Partial functional correction of xeroderma pigmentosum group A cells by suppressor tRNA. Hum. Gene Ther. 10, 2209–2219 (1999).
Wang, J. et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 604, 343–348 (2022). The study shows the first in vivo administration of sup-tRNAs as AAV formulations for episomal expression and presents the efficacy in different mouse tissues.
Albers, S. et al. Repurposing tRNAs for nonsense suppression. Nat. Commun. 12, 3850 (2021).
Albers, S. et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 618, 842–848 (2023). This publication presents improved sup-tRNA designs for in vivo LNP administrations to target liver and lung and demonstrates a clinical benefit for a sup-tRNAArg.
Sako, Y., Usuki, F. & Suga, H. A novel therapeutic approach for genetic diseases by introduction of suppressor tRNA. Nucleic Acids Symp. Ser. 50, 239–240 (2006).
Cervettini, D. et al. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase-tRNA pairs. Nat. Biotechnol. 38, 989–999 (2020).
Fan, C., Xiong, H., Reynolds, N. M. & Soll, D. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 43, e156 (2015).
Jewel, D. et al. Virus-assisted directed evolution of enhanced suppressor tRNAs in mammalian cells. Nat. Methods 20, 95–103 (2023).
Katoh, T., Iwane, Y. & Suga, H. Logical engineering of D-arm and T-stem of tRNA that enhances d-amino acid incorporation. Nucleic Acids Res. 45, 12601–12610 (2017).
Prabhakar, A. et al. Uncovering translation roadblocks during the development of a synthetic tRNA. Nucleic Acids Res. 50, 10201–10211 (2022).
Wang, L. & Schultz, P. G. A general approach for the generation of orthogonal tRNAs. Chem. Biol. 8, 883–890 (2001).
Giege, R. & Eriani, G. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 51, 1528–1570 (2023).
Burgess, R. W. & Storkebaum, E. tRNA dysregulation in neurodevelopmental and neurodegenerative diseases. Annu. Rev. Cell Dev. Biol. 39, 223–252 (2023).
Ishimura, R. et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 (2014).
Orellana, E. A., Siegal, E. & Gregory, R. I. tRNA dysregulation and disease. Nat. Rev. Genet. 23, 651–664 (2022).
Spaulding, E. L. et al. The integrated stress response contributes to tRNA synthetase-associated peripheral neuropathy. Science 373, 1156–1161 (2021). The study shows that ribosome stalling owing to shortage of the cognate tRNA activates the integrated stress response.
Zuko, A. et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 373, 1161–1166 (2021). This study, published in the same issue of the journal as ref. 47, reports that tRNA sequestration by the cognate AARS results in ribosome stalling and selects tRNA supplementation as a therapeutic modality.
Masvidal, L. et al. Assessing the residual CFTR gene expression in human nasal epithelium cells bearing CFTR splicing mutations causing cystic fibrosis. Eur. J. Hum. Genet. 22, 784–791 (2014).
McCague, A. F. et al. Correlating cystic fibrosis transmembrane conductance regulator function with clinical features to inform precision treatment of cystic fibrosis. Am. J. Respir. Crit. Care Med. 199, 1116–1126 (2019).
Tan, K., Stupack, D. G. & Wilkinson, M. F. Nonsense-mediated RNA decay: an emerging modulator of malignancy. Nat. Rev. Cancer 22, 437–451 (2022).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Rice, A. M. & McLysaght, A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 8, 14366 (2017).
Sandweiss, A. J., Brandt, V. L. & Zoghbi, H. Y. Advances in understanding of Rett syndrome and MECP2 duplication syndrome: prospects for future therapies. Lancet Neurol. 19, 689–698 (2020).
Grosjean, H. & Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44, 8020–8040 (2016).
Westhof, E., Thornlow, B., Chan, P. P. & Lowe, T. M. Eukaryotic tRNA sequences present conserved and amino acid-specific structural signatures. Nucleic Acids Res. 50, 4100–4112 (2022). The study presents evidence for tRNA sequence motifs that are anticodon-specific and conserved across tRNAs and, thus, could be instructive on sup-tRNA designs.
Yang, Y. M. et al. Comprehensive genetic diagnosis of patients with Duchenne/Becker muscular dystrophy (DMD/BMD) and pathogenicity analysis of splice site variants in the DMD gene. J. Zhejiang Univ. Sci. B 20, 753–765 (2019).
Duan, D., Goemans, N., Takeda, S., Mercuri, E. & Aartsma-Rus, A. Duchenne muscular dystrophy. Nat. Rev. Dis. Prim. 7, 13 (2021).
Veit, G. et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol. Biol. Cell 27, 424–433 (2016).
Xue, X. et al. Identification of the amino acids inserted during suppression of CFTR nonsense mutations and determination of their functional consequences. Hum. Mol. Genet. 26, 3116–3129 (2017).
Zaher, A., ElSaygh, J., Elsori, D., ElSaygh, H. & Sanni, A. A review of Trikafta: triple cystic fibrosis transmembrane conductance regulator (CFTR) modulator therapy. Cureus 13, e16144 (2021).
Pareyson, D., Scaioli, V. & Laura, M. Clinical and electrophysiological aspects of Charcot-Marie-Tooth disease. Neuromol. Med. 8, 3–22 (2006).
Niehues, S. et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 6, 7520 (2015).
Nangle, L. A., Zhang, W., Xie, W., Yang, X. L. & Schimmel, P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl Acad. Sci. USA 104, 11239–11244 (2007).
Kuzmin, D. A. et al. The clinical landscape for AAV gene therapies. Nat. Rev. Drug Discov. 20, 173–174 (2021).
Ma, H. et al. Pol III promoters to express small RNAs: delineation of transcription initiation. Mol. Ther. Nucleic Acids 3, e161 (2014).
Gao, Z., Herrera-Carrillo, E. & Berkhout, B. RNA polymerase II activity of type 3 pol III promoters. Mol. Ther. Nucleic Acids 12, 135–145 (2018).
Traboni, C., Ciliberto, G. & Cortese, R. Mutations in box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell 36, 179–187 (1984).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Johnson, P. F. & Abelson, J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature 302, 681–687 (1983).
Phizicky, E. M. & Hopper, A. K. tRNA biology charges to the front. Genes Dev. 24, 1832–1860 (2010).
van Tol, H. & Beier, H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res. 16, 1951–1966 (1988).
Ogawa, A., Doi, Y. & Matsushita, N. Improvement of in vitro-transcribed amber suppressor tRNAs toward higher suppression efficiency in wheat germ extract. Org. Biomol. Chem. 9, 8495–8503 (2011).
Walker, S. E. & Fredrick, K. Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 (2008).
Amrani, N. et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118 (2004).
Achsel, T. & Gross, H. J. Identity determinants of human tRNA(Ser): sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 12, 3333–3338 (1993).
Breitschopf, K., Achsel, T., Busch, K. & Gross, H. J. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 23, 3633–3637 (1995).
Tsunoda, M. et al. Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms. Nucleic Acids Res. 35, 4289–4300 (2007).
Shen, N., Guo, L., Yang, B., Jin, Y. & Ding, J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 34, 3246–3258 (2006).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Manickam, N., Joshi, K., Bhatt, M. J. & Farabaugh, P. J. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 44, 1871–1881 (2016).
Nguyen, H. A., Hoffer, E. D. & Dunham, C. M. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGProfor decoding. J. Biol. Chem. 294, 5281–5291 (2019).
Giege, R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 15, 1007–1014 (2008).
Ledoux, S. & Uhlenbeck, O. C. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell 31, 114–123 (2008).
LaRiviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 (2001).
Nguyen, H. A., Sunita, S. & Dunham, C. M. Disruption of evolutionarily correlated tRNA elements impairs accurate decoding. Proc. Natl Acad. Sci. USA 117, 16333–16338 (2020).
Schrader, J. M., Chapman, S. J. & Uhlenbeck, O. C. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl Acad. Sci. USA 108, 5215–5220 (2011).
Yarus, M., Cline, S., Raftery, L., Wier, P. & Bradley, D. The translational efficiency of tRNA is a property of the anticodon arm. J. Biol. Chem. 261, 10496–10505 (1986).
Uhlenbeck, O. C. & Schrader, J. M. Evolutionary tuning impacts the design of bacterial tRNAs for the incorporation of unnatural amino acids by ribosomes. Curr. Opin. Chem. Biol. 46, 138–145 (2018).
Wangen, J. R. & Green, R. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife 9, e52611 (2020).
Kachale, A. et al. Short tRNA anticodon stem and mutant eRF1 allow stop codon reassignment. Nature 613, 751–758 (2023).
Hilal, T. et al. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science 376, 1338–1343 (2022).
Serrao, V. H. B. et al. The unique tRNA(Sec) and its role in selenocysteine biosynthesis. Amino Acids 50, 1145–1167 (2018).
Ogawa, A., Hayami, M., Sando, S. & Aoyama, Y. A concept for selection of codon-suppressor tRNAs based on read-through ribosome display in an in vitro compartmentalized cell-free translation system. J. Nucleic Acids 2012, 538129 (2012).
Keeling, K. M. et al. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10, 691–703 (2004).
Bowie, A. G. & Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8, 911–922 (2008).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Nelson, J. et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 6, eaaz6893 (2020).
Agris, P. F. The importance of being modified: an unrealized code to RNA structure and function. RNA 21, 552–554 (2015).
Kierzek, E. & Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480 (2003).
Vare, V. Y., Eruysal, E. R., Narendran, A., Sarachan, K. L. & Agris, P. F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 7, 29 (2017).
Jockel, S. et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 (2012).
Rimbach, K., Kaiser, S., Helm, M., Dalpke, A. H. & Eigenbrod, T. 2′-O-Methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J. Innate Immun. 7, 482–493 (2015).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Colella, P., Ronzitti, G. & Mingozzi, F. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104 (2018).
Godbout, K. & Tremblay, J. P. Delivery of RNAs to specific organs by lipid nanoparticles for gene therapy. Pharmaceutics 14, 2129 (2022).
Hald Albertsen, C. et al. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 188, 114416 (2022).
Herrmann, I. K., Wood, M. J. A. & Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 16, 748–759 (2021).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Li, C. & Samulski, R. J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 21, 255–272 (2020).
Ling, Q., Herstine, J. A., Bradbury, A. & Gray, S. J. AAV-based in vivo gene therapy for neurological disorders. Nat. Rev. Drug Discov. 22, 789–806 (2023). The paper reviews the state-of-the-art of engineered AAV delivery systems with optimized safety and efficacy to treat neurological disorders that are applicable to tRNA medicines.
Patel, S., Ryals, R. C., Weller, K. K., Pennesi, M. E. & Sahay, G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release 303, 91–100 (2019).
Paunovska, K., Loughrey, D. & Dahlman, J. E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265–280 (2022). The review presents the state-of-the-art of non-viral delivery systems for RNA therapeutics, many of which are applicable to tRNA therapeutics.
Pupo, A. et al. AAV vectors: the Rubik’s cube of human gene therapy. Mol. Ther. 30, 3515–3541 (2022).
Sahin, U., Kariko, K. & Tureci, O. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Zhang, J., Shrivastava, S., Cleveland, R. O. & Rabbitts, T. H. Lipid-mRNA nanoparticle designed to enhance intracellular delivery mediated by shock waves. ACS Appl. Mater. Interfaces 11, 10481–10491 (2019).
Zhang, Y. et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnol. 20, 279 (2022).
Gaudet, D. et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther. 20, 361–369 (2013).
Frohlich, D. et al. Dual-function AAV gene therapy reverses late-stage Canavan disease pathology in mice. Front. Mol. Neurosci. 15, 1061257 (2022).
Sumner, C. J. & Crawford, T. O. Two breakthrough gene-targeted treatments for spinal muscular atrophy: challenges remain. J. Clin. Invest. 128, 3219–3227 (2018).
Nieuwenhuis, B. et al. Improving adeno-associated viral (AAV) vector-mediated transgene expression in retinal ganglion cells: comparison of five promoters. Gene Ther. 30, 503–519 (2023).
Meyer-Berg, H. et al. Identification of AAV serotypes for lung gene therapy in human embryonic stem cell-derived lung organoids. Stem Cell Res. Ther. 11, 448 (2020).
Palaschak, B., Herzog, R. W. & Markusic, D. M. AAV-mediated gene delivery to the liver: overview of current technologies and methods. Methods Mol. Biol. 1950, 333–360 (2019).
Gernoux, G. et al. Muscle-directed delivery of an AAV1 vector leads to capsid-specific T cell exhaustion in nonhuman primates and humans. Mol. Ther. 28, 747–757 (2020).
Valdmanis, P. N. & Kay, M. A. Future of rAAV gene therapy: platform for RNAi, gene editing, and beyond. Hum. Gene Ther. 28, 361–372 (2017).
Srivastava, A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 21, 75–80 (2016).
Lisowski, L. et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506, 382–386 (2014).
George, L. A. et al. Multiyear factor VIII expression after AAV gene transfer for hemophilia A. N. Engl. J. Med. 385, 1961–1973 (2021).
Gonzalez-Sandoval, A. et al. The AAV capsid can influence the epigenetic marking of rAAV delivered episomal genomes in a species dependent manner. Nat. Commun. 14, 2448 (2023).
Dobrowolski, C., Paunovska, K., Hatit, M. Z. C., Lokugamage, M. P. & Dahlman, J. E. Therapeutic RNA delivery for COVID and other diseases. Adv. Healthc. Mater. 10, e2002022 (2021).
Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018).
Kauffman, K. J., Webber, M. J. & Anderson, D. G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release 240, 227–234 (2016).
Xia, Y., Tian, J. & Chen, X. Effect of surface properties on liposomal siRNA delivery. Biomaterials 79, 56–68 (2016).
Kulkarni, J. A. et al. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 16, 630–643 (2021). This review summarizes the developments of delivery systems for clinical translation of antisense oligonucleotide-based therapy, which owing to the similar cargo size would be applicable to tRNA medicines.
Maier, M. A. et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 21, 1570–1578 (2013).
Sabnis, S. et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
Jiang, L. et al. Systemic messenger RNA as an etiological treatment for acute intermittent porphyria. Nat. Med. 24, 1899–1909 (2018).
Hewitt, S. L. et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36gamma, and OX40L mRNAs. Sci. Transl. Med. 11, eaat9143 (2019).
Kulkarni, J. A., Cullis, P. R. & van der Meel, R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 28, 146–157 (2018).
Cheng, X. & Lee, R. J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 99, 129–137 (2016).
Suk, J. S., Xu, Q., Kim, N., Hanes, J. & Ensign, L. M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 99, 28–51 (2016).
Paunovska, K. et al. Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery. ACS Nano 12, 8341–8349 (2018).
Eygeris, Y., Patel, S., Jozic, A. & Sahay, G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 20, 4543–4549 (2020).
Pardridge, W. M. Brain gene therapy with Trojan horse lipid nanoparticles. Trends Mol. Med. 29, 343–353 (2023).
Riley, R. S. et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv. 7, eaba1028 (2021).
Zhang, H., Leal, J., Soto, M. R., Smyth, H. D. C. & Ghosh, D. Aerosolizable lipid nanoparticles for pulmonary delivery of mRNA through design of experiments. Pharmaceutics 12, 1042 (2020).
Devoldere, J. et al. Non-viral delivery of chemically modified mRNA to the retina: subretinal versus intravitreal administration. J. Control. Release 307, 315–330 (2019).
Nabhan, J. F. et al. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 6, 20019 (2016).
Turnbull, I. C. et al. Myocardial delivery of lipidoid nanoparticle carrying modRNA induces rapid and transient expression. Mol. Ther. 24, 66–75 (2016).
Dilliard, S. A., Cheng, Q. & Siegwart, D. J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl Acad. Sci. USA 118, e2109256118 (2021).
Sheridan, C. Why gene therapies must go virus-free. Nat. Biotechnol. 41, 737–739 (2023).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Ma, F. et al. Neurotransmitter-derived lipidoids (NT-lipidoids) for enhanced brain delivery through intravenous injection. Sci. Adv. 6, eabb4429 (2020).
Shi, N., Zhang, Y., Zhu, C., Boado, R. J. & Pardridge, W. M. Brain-specific expression of an exogenous gene after i.v. administration. Proc. Natl Acad. Sci. USA 98, 12754–12759 (2001).
Zhang, Y., Schlachetzki, F. & Pardridge, W. M. Global non-viral gene transfer to the primate brain following intravenous administration. Mol. Ther. 7, 11–18 (2003).
Zhang, Y. F., Boado, R. J. & Pardridge, W. M. Absence of toxicity of chronic weekly intravenous gene therapy with pegylated immunoliposomes. Pharm. Res. 20, 1779–1785 (2003).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Muthu, S., Bapat, A., Jain, R., Jeyaraman, N. & Jeyaraman, M. Exosomal therapy — a new frontier in regenerative medicine. Stem Cell Investig. 8, 7 (2021).
Dixson, A. C., Dawson, T. R., Di Vizio, D. & Weaver, A. M. Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 24, 454–476 (2023).
Rezaie, J., Feghhi, M. & Etemadi, T. A review on exosomes application in clinical trials: perspective, questions, and challenges. Cell Commun. Signal. 20, 145 (2022).
Yang, Y., Hong, Y., Cho, E., Kim, G. B. & Kim, I. S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 7, 1440131 (2018).
Bunggulawa, E. J. et al. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 16, 81 (2018).
Jeyaraman, M. et al. Mesenchymal stem cell-derived exosomes: a potential therapeutic avenue in knee osteoarthritis. Cartilage 13, 1572S–1585S (2021).
Cho, E. et al. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. J. Control. Release 279, 326–335 (2018).
Kamerkar, S. et al. Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv. 8, eabj7002 (2022).
Yokoi, A. et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 5, eaax8849 (2019).
Katakowski, M. et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 335, 201–204 (2013).
Yang, T. et al. Delivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell-secreted exosome nanovesicles for the treatment of brain cancer. AAPS J. 19, 475–486 (2017).
Mendt, M. et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 3, e99263 (2018).
Zhu, X. et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles 6, 1324730 (2017).
Kojima, R. et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 9, 1305 (2018).
Liu, C. & Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 9, 1015–1028 (2019).
Guan, S. et al. Self-assembled peptide-poloxamine nanoparticles enable in vitro and in vivo genome restoration for cystic fibrosis. Nat. Nanotechnol. 14, 287–297 (2019).
Cooney, A. L., Singh, B. K. & Sinn, P. L. Hybrid nonviral/viral vector systems for improved piggyBac DNA transposon in vivo delivery. Mol. Ther. 23, 667–674 (2015).
Arribere, J. A. et al. Translation readthrough mitigation. Nature 534, 719–723 (2016). The publication presents evidence for spontaneous readthrough at natural termination codons and activation of surveillance mechanisms to clear readthrough proteins, which should be considered in the context of safe tRNA therapeutics.
Muller, M. B. D., Kasturi, P., Jayaraj, G. G. & Hartl, F. U. Mechanisms of readthrough mitigation reveal principles of GCN1-mediated translational quality control. Cell 186, 3227–3244 (2023).
Sitron, C. S. & Brandman, O. Detection and degradation of stalled nascent chains via ribosome-associated quality control. Annu. Rev. Biochem. 89, 417–442 (2020).
Loughran, G. et al. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938 (2014).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Swart, E. C., Serra, V., Petroni, G. & Nowacki, M. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell 166, 691–702 (2016).
Pavon-Eternod, M., Gomes, S., Rosner, M. R. & Pan, T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19, 461–466 (2013).
Hughes, L. A. et al. Copy number variation in tRNA isodecoder genes impairs mammalian development and balanced translation. Nat. Commun. 14, 2210 (2023).
Rosa, S. S., Prazeres, D. M. F., Azevedo, A. M. & Marques, M. P. C. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine 39, 2190–2200 (2021).
Swayze, E. E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007).
Food and Drug Administration (FDA) Cellular, Tissue, and Gene Therapies Advisory Committee. Toxicity risks of adeno-associated virus (AAV) vectors for gene therapy (GT). https://www.fda.gov/media/151599/download (2021).
Ertl, H. C. J. Immunogenicity and toxicity of AAV gene therapy. Front. Immunol. 13, 975803 (2022).
Boutin, S. et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 21, 704–712 (2010).
Calcedo, R., Vandenberghe, L. H., Gao, G., Lin, J. & Wilson, J. M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 199, 381–390 (2009).
Manno, C. S. et al. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 12, 342–347 (2006).
Meliani, A. et al. Antigen-selective modulation of AAV immunogenicity with tolerogenic rapamycin nanoparticles enables successful vector re-administration. Nat. Commun. 9, 4098 (2018).
Jordan, S. C. et al. IgG endopeptidase in highly sensitized patients undergoing transplantation. N. Engl. J. Med. 377, 442–453 (2017).
Leborgne, C. et al. IgG-cleaving endopeptidase enables in vivo gene therapy in the presence of anti-AAV neutralizing antibodies. Nat. Med. 26, 1096–1101 (2020).
Smith, R. H. Adeno-associated virus integration: virus versus vector. Gene Ther. 15, 817–822 (2008).
Nault, J. C. et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 47, 1187–1193 (2015).
Berns, K. I. & Muzyczka, N. AAV: an overview of unanswered questions. Hum. Gene Ther. 28, 308–313 (2017).
Kedmi, R., Ben-Arie, N. & Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 31, 6867–6875 (2010).
Lv, H., Zhang, S., Wang, B., Cui, S. & Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 114, 100–109 (2006).
Kozma, G. T., Shimizu, T., Ishida, T. & Szebeni, J. Anti-PEG antibodies: properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Adv. Drug Deliv. Rev. 154–155, 163–175 (2020).
Mingozzi, F. & High, K. A. Overcoming the host immune response to adeno-associated virus gene delivery vectors: the race between clearance, tolerance, neutralization, and escape. Annu. Rev. Virol. 4, 511–534 (2017).
Pei, D. & Buyanova, M. Overcoming endosomal entrapment in drug delivery. Bioconjug. Chem. 30, 273–283 (2019).
Choe, B. K. & Taylor, M. W. Kinetics of synthesis and characterization of transfer RNA precursors in mammalian cells. Biochim. Biophys. Acta 272, 275–287 (1972).
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018).
Sago, C. D. et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl Acad. Sci. USA 115, E9944–E9952 (2018).
Linde, L. et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest. 117, 683–692 (2007).
Behrens, A., Rodschinka, G. & Nedialkova, D. D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815 (2021).
Pinkard, O., McFarland, S., Sweet, T. & Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11, 4104 (2020).
Shigematsu, M. et al. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res. 45, e70 (2017).
Gogakos, T. et al. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Shi, J. et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 23, 424–436 (2021).
Honda, S., Shigematsu, M., Morichika, K., Telonis, A. G. & Kirino, Y. Four-leaf clover qRT-PCR: a convenient method for selective quantification of mature tRNA. RNA Biol. 12, 501–508 (2015).
Erber, L. et al. LOTTE-seq (long hairpin oligonucleotide based tRNA high-throughput sequencing): specific selection of tRNAs with 3′-CCA end for high-throughput sequencing. RNA Biol. 17, 23–32 (2020).
Nagai, A., Mori, K., Shiomi, Y. & Yoshihisa, T. OTTER, a new method quantifying absolute amounts of tRNAs. RNA 27, 628–640 (2021).
Tsukamoto, Y., Nakamura, Y., Hirata, M., Sakate, R. & Kimura, T. i-tRAP (individual tRNA acylation PCR): a convenient method for selective quantification of tRNA charging. RNA 29, 111–122 (2022).
Thomas, N. K. et al. Direct nanopore sequencing of individual full length tRNA strands. ACS Nano 15, 16642–16653 (2021).
Lucas, M. C. et al. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. (in the press). The publication presents a nanopore-based tRNA sequencing method to determine tRNA abundance and modification pattern in a single sequencing run.
Shi, Y., Inoue, H., Wu, J. C. & Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115–130 (2017).
Tricot, T., Verfaillie, C. M. & Kumar, M. Current status and challenges of human induced pluripotent stem cell-derived liver models in drug discovery. Cells 11, 442 (2022).
Dekkers, J. F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013).
Han, S. T. et al. Residual function of cystic fibrosis mutants predicts response to small molecule CFTR modulators. JCI Insight 3, e121159 (2018).
Silva, I. A. L., Laselva, O. & Lopes-Pacheco, M. Advances in preclinical in vitro models for the translation of precision medicine for cystic fibrosis. J. Pers. Med. 12, 1321 (2022).
Piga, D. et al. Human induced pluripotent stem cell models for the study and treatment of Duchenne and Becker muscular dystrophies. Ther. Adv. Neurol. Disord. 12, 1756286419833478 (2019).
Uchimura, T., Asano, T., Nakata, T., Hotta, A. & Sakurai, H. A muscle fatigue-like contractile decline was recapitulated using skeletal myotubes from Duchenne muscular dystrophy patient-derived iPSCs. Cell Rep. Med. 2, 100298 (2021).
Wadman, M. FDA no longer has to require animal testing for new drugs. Science 379, 127–128 (2023).
Crooke, S. T. Meeting the needs of patients with ultrarare diseases. Trends Mol. Med. 28, 87–96 (2022).
May, M. Rare-disease researchers pioneer a unique approach to clinical trials. Nat. Med. 29, 1884–1886 (2023).
Ornes, S. Core concept: basket trial approach capitalizes on the molecular mechanisms of tumors. Proc. Natl Acad. Sci. USA 113, 7007–7008 (2016).
Park, J. J. H., Hsu, G., Siden, E. G., Thorlund, K. & Mills, E. J. An overview of precision oncology basket and umbrella trials for clinicians. CA Cancer J. Clin. 70, 125–137 (2020).
Ko, W., Porter, J. J., Sipple, M. T., Edwards, K. M. & Lueck, J. D. Efficient suppression of endogenous CFTR nonsense mutations using anticodon-engineered transfer RNAs. Mol. Ther. Nucleic Acids 28, 685–701 (2022).
Hoagland, M. B., Stephenson, M. L., Scott, J. F., Hecht, L. I. & Zamecnik, P. C. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 231, 241–257 (1958).
Person, S. & Osborn, M. The conversion of amber suppressors to ochre suppressors. Proc. Natl Acad. Sci. USA 60, 1030–1037 (1968).
Langer, R. & Folkman, J. Polymers for the sustained release of proteins and other macromolecules. Nature 263, 797–800 (1976).
Lee, B. J., Worland, P. J., Davis, J. N., Stadtman, T. C. & Hatfield, D. L. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem. 264, 9724–9727 (1989).
Roehr, B. Fomivirsen approved for CMV retinitis. J. Int. Assoc. Physicians AIDS Care 4, 14–16 (1998).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Biela, A. et al. The diverse structural modes of tRNA binding and recognition. J. Biol. Chem. 299, 104966 (2023).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Acknowledgements
We acknowledge the helpful discussions with H.-Q. Mao (Institute for NanoBioTechnology, Johns Hopkins University), and P. Kaplan and P. Eimon (Tevard Biosciences). Work in the Coller Lab is supported by NIH grant (R35GM144114), Bloomberg Philanthropies and the Syngap Research Fund, and the research of Z.I. is supported by grants of the Deutsche Forschungsgemeinschaft DFG (IG 73/21-1), NIH (1R01HL136414-05) and Cystic Fibrosis Foundation (IGNATO2010).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
Z.I. is co-inventor on several licensed and non-licensed patents related to the use of tRNAs as therapeutics and sup-tRNA design and is a member of the scientific advisory board of Tevard Biosciences. J.C. is scientific co-founder of Tevard Biosciences and a member of their scientific advisory board.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Adeno-associated virus
-
(AAV). Non-pathogenic, small single-stranded DNA virus, whose genome (4.7 kb) encodes four non-structural rep proteins, three capsid (cap) proteins and assembly-activating protein, flanked by two AAV-specific palindromic ITRs (145 bp).
- Aminoacyl-tRNA synthetase
-
(AARS). A universal enzyme family that aminoacylates tRNAs with their cognate amino acid.
- Basket trials
-
Also known as bucket trials; a type of clinical trial for patients with different diseases with the same mutation or biomarker.
- Episomal expression
-
A non-integrated extrachromosomal circular DNA from a viral genome that replicates and is transcribed independently in the eukaryotic nucleus.
- Missense mutation
-
A genetic alteration within the protein-coding sequence leading to a change of the encoded amino acid which may alter the function of a protein.
- Natural suppressor tRNAs
-
Native suppressors of nonsense mutations (mostly in bacteria and yeast) arise from mutation in sense codon decoding tRNA genes enabling the mutant tRNA to translate a stop codon.
- Nonsense-mediated mRNA decay
-
(NMD). A pathway that rapidly degrades mRNA in response to nonsense mutations, which can arise owing to errors in transcription or failure to remove intronic regions, altering the natural reading frame.
- Nonsense mutation
-
A genetic alteration within the protein-coding sequence exchanging a sense codon (that is, encoding an amino acid) to a termination or stop codon, resulting in a loss of protein function.
- Recombinant AAV
-
(rAAV). Engineered AAV, in which the DNA of interest replaces the viral sequences encoding for rep and cap genes, whereas both cis-packaging ITR signals are retained.
- Termination codons
-
Also known as stop codons. Three codons, named amber (UAG), ochre (UAA) and opal (UGA), terminate mRNA translation and, through pairing with the release factor (eRF1 in eukaryotes), release the newly synthesized protein.
- Therapeutic threshold
-
The therapeutic threshold, which is established for each disease based on calculations, descriptive methods and clinical practice, describes the probability of disease at which the condition between treatment and no treatment is the same.
- Toll-like receptors
-
Mediators of inflammatory pathways mediating the immune response towards a variety of pathogen-derived ligands, including DNA, double-stranded RNA, single-stranded RNA and oligonucleotides.
- tRNA isoacceptor family
-
A group of all tRNA isoacceptors carrying the same amino acid.
- tRNA isoacceptors
-
Different tRNA species, which are aminoacylated with the same amino acid, but differ in their anticodon sequences.
- tRNA isodecoders
-
tRNA species that are aminoacylated with the same amino acids and bear the same anticodon, but differ elsewhere in their sequences.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Coller, J., Ignatova, Z. tRNA therapeutics for genetic diseases. Nat Rev Drug Discov 23, 108–125 (2024). https://doi.org/10.1038/s41573-023-00829-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-023-00829-9