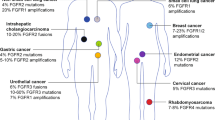
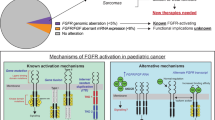
Abstract
Fibroblast growth factor (FGF) signalling via FGF receptors (FGFR1–4) orchestrates fetal development and contributes to tissue and whole-body homeostasis, but can also promote tumorigenesis. Various agents, including pan-FGFR inhibitors (erdafitinib and futibatinib), FGFR1/2/3 inhibitors (infigratinib and pemigatinib), as well as a range of more-specific agents, have been developed and several have entered clinical use. Erdafitinib is approved for patients with urothelial carcinoma harbouring FGFR2/3 alterations, and futibatinib and pemigatinib are approved for patients with cholangiocarcinoma harbouring FGFR2 fusions and/or rearrangements. Clinical benefit from these agents is in part limited by hyperphosphataemia owing to off-target inhibition of FGFR1 as well as the emergence of resistance mutations in FGFR genes, activation of bypass signalling pathways, concurrent TP53 alterations and possibly epithelial–mesenchymal transition-related isoform switching. The next generation of small-molecule inhibitors, such as lirafugratinib and LOXO-435, and the FGFR2-specific antibody bemarituzumab are expected to have a reduced risk of hyperphosphataemia and the ability to overcome certain resistance mutations. In this Review, we describe the development and current clinical role of FGFR inhibitors and provide perspective on future research directions including expansion of the therapeutic indications for use of FGFR inhibitors, combination of these agents with immune-checkpoint inhibitors and the application of novel technologies, such as artificial intelligence.
Key points
-
Several small-molecule pan-fibroblast growth factor receptor (FGFR) inhibitors (such as erdafitinib and futibatinib) and FGFR1–3 inhibitors (such as pemigatinib and infigratinib) have been approved for the treatment of patients with various cancers that harbour FGFR alterations detected using their companion diagnostics.
-
Objective response rates of patients with solid tumours receiving approved FGFR inhibitors are ~20–40%, whereas patients with FGFR1-rearranged myeloid or lymphoid neoplasms receiving pemigatinib have a complete response rate of 77%.
-
Gatekeeper, molecular-brake and other secondary FGFR alterations, activation of bypass signalling and TP53 alterations can all lead to resistance to FGFR inhibitors, and hyperphosphataemia owing to off-tumour inhibition of FGFR1 can lead to dose reduction or treatment discontinuation.
-
Prospective data indicate that the activity of selective FGFR2 or FGFR3 inhibitors is unaffected by gatekeeper mutations and that these agents are less likely to induce hyperphosphataemia; these more-selective agents are expected to form the third generation of FGFR inhibitors for patients with FGFR2/3-altered cancers.
-
The anti-FGFR2b antibody bemarituzumab is currently being tested in combination with chemotherapy in phase III trials in patients with gastric or gastro-oesophageal junction adenocarcinoma harbouring FGFR2 amplifications and/or overexpression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Katoh, M. & Nakagama, H. FGF receptors: cancer biology and therapeutics. Med. Res. Rev. 34, 280–300 (2014).
Babina, I. S. & Turner, N. C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332 (2017).
Krook, M. A. et al. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer 124, 880–892 (2021).
Hung, K. L. et al. Targeted profiling of human extrachromosomal DNA by CRISPR–CATCH. Nat. Genet. 54, 1746–1754 (2022).
Singh, D. et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 (2012).
Wu, Y. M. et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 3, 636–647 (2013).
Reeser, J. W. et al. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J. Mol. Diagn. 19, 682–696 (2017).
Jain, P. et al. Novel FGFR2-INA fusion identified in two low-grade mixed neuronal-glial tumors drives oncogenesis via MAPK and PI3K/mTOR pathway activation. Acta Neuropathol. 136, 167–169 (2018).
Qin, A. et al. Detection of known and novel FGFR fusions in non-small cell lung cancer by comprehensive genomic profiling. J. Thorac. Oncol. 14, 54–62 (2019).
Liu, Y. J. et al. Calcified chondroid mesenchymal neoplasms with FN1-receptor tyrosine kinase gene fusions including FGFR2, FGFR1, MERTK, NTRK1, and TEK: a molecular and clinicopathologic analysis. Mod. Pathol. 34, 1373–1383 (2021).
Gu, W. et al. Comprehensive identification of FGFR1-4 alterations in 5557 Chinese patients with solid tumors by next-generation sequencing. Am. J. Cancer Res. 11, 3893–3906 (2021).
Tao, Z. et al. Profiling receptor tyrosine kinase fusions in chinese breast cancers. Front. Oncol. 11, 741142 (2021).
Georgantzoglou, N., Shen, G., Jour, G. & Linos, K. A case of FN1-fused calcified chondroid mesenchymal neoplasm of the hand with novel FGFR3 partner gene. Genes Chromosomes Cancer 62, 237–241 (2023).
Katoh, M. & Katoh, M. Precision medicine for human cancers with Notch signaling dysregulation. Int. J. Mol. Med. 45, 279–297 (2020).
Helsten, T. et al. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin. Cancer Res. 22, 259–267 (2016).
Sun, Y. et al. A comprehensive pan-cancer study of fibroblast growth factor receptor aberrations in Chinese cancer patients. Ann. Transl. Med. 8, 1290 (2020).
Katoh, M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 16, 105–122 (2019).
Katoh, M. FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis. Int. J. Mol. Med. 38, 3–15 (2016).
Porta, C., Giglione, P., Liguigli, W. & Paglino, C. Dovitinib (CHIR258, TKI258): structure, development and preclinical and clinical activity. Future Oncol. 11, 39–50 (2015).
Hilberg, F. et al. Triple angiokinase inhibitor nintedanib directly inhibits tumor cell growth and induces tumor shrinkage via blocking oncogenic receptor tyrosine kinases. J. Pharmacol. Exp. Ther. 364, 494–503 (2018).
Loriot, Y. et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 381, 338–348 (2019).
Goyal, L. et al. Futibatinib for FGFR2-rearranged intrahepatic cholangiocarcinoma. N. Engl. J. Med. 388, 228–239 (2023).
Abou-Alfa, G. K. et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 21, 671–684 (2020).
Javle, M. et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol. Hepatol. 6, 803–815 (2021).
Gotlib, J. et al. A phase 2 study of pemigatinib (FIGHT-203; INCB054828) in patients with myeloid/lymphoid neoplasms (MLNs) with fibroblast growth factor receptor 1 (FGFR1) rearrangement. Blood 138, 385 (2021).
Katoh, M. WNT and FGF gene clusters. Int. J. Oncol. 21, 1269–1273 (2002).
Ornitz, D. M. & Itoh, N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015).
Li, X. The FGF metabolic axis. Front. Med. 13, 511–530 (2019).
Eswarakumar, V. P., Lax, I. & Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor. Rev. 16, 139–149 (2005).
Katoh, M. Therapeutics targeting FGF signaling network in human diseases. Trends Pharmacol. Sci. 37, 1081–1096 (2016).
Chen, L. et al. Structural basis for FGF hormone signalling. Nature 618, 862–870 (2023).
Chen, H. et al. Elucidation of a four-site allosteric network in fibroblast growth factor receptor tyrosine kinases. eLife 6, e21137 (2017).
Marsiglia, W. M. et al. A conserved allosteric pathway in tyrosine kinase regulation. Structure 27, 1308–1315 (2019).
Chioni, A. M. & Grose, R. P. Biological significance and targeting of the FGFR axis in cancer. Cancers 13, 5681 (2021).
Wong, A., Lamothe, B., Lee, A., Schlessinger, J. & Lax, I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc. Natl Acad. Sci. USA 99, 6684–6689 (2002).
Haugsten, E. M., Malecki, J., Bjørklund, S. M., Olsnes, S. & Wesche, J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 19, 3390–3403 (2008).
Guo, C. et al. Sprouty 2 disturbs FGFR3 degradation in thanatophoric dysplasia type II: a severe form of human achondroplasia. Cell Signal. 20, 1471–1477 (2008).
Porębska, N. et al. Targeting cellular trafficking of fibroblast growth factor receptors as a strategy for selective cancer treatment. J. Clin. Med. 8, 7 (2018).
Cullen, P. J. & Steinberg, F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018).
Dai, S., Zhou, Z., Chen, Z., Xu, G. & Chen, Y. Fibroblast growth factor receptors (FGFRs): structures and small molecule inhibitors. Cells 8, 614 (2019).
Rouanne, M. et al. Novel therapeutic targets in advanced urothelial carcinoma. Crit. Rev. Oncol. Hematol. 98, 106–115 (2016).
Bahleda, R. et al. Multicenter phase I study of erdafitinib (JNJ-42756493), oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced or refractory solid tumors. Clin. Cancer Res. 25, 4888–4897 (2019).
Patel, V. G., Oh, W. K. & Galsky, M. D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J. Clin. 70, 404–423 (2020).
Ardizzone, A. et al. Role of fibroblast growth factors receptors (FGFRs) in brain tumors, focus on astrocytoma and glioblastoma. Cancers 12, 3825 (2020).
Georgescu, M. M., Islam, M. Z., Li, Y., Traylor, J. & Nanda, A. Novel targetable FGFR2 and FGFR3 alterations in glioblastoma associate with aggressive phenotype and distinct gene expression programs. Acta Neuropathol. Commun. 9, 69 (2021).
Schittenhelm, J. et al. FGFR3 overexpression is a useful detection tool for FGFR3 fusions and sequence variations in glioma. Neurooncol. Pract. 8, 209–221 (2021).
Sobhani, N., Fan, C., Flores-Villanueva, P. O., Generali, D. & Li, Y. The fibroblast growth factor receptors in breast cancer: from oncogenesis to better treatments. Int. J. Mol. Sci. 21, 2011 (2020).
Santolla, M. F. & Maggiolini, M. The FGF/FGFR system in breast cancer: oncogenic features and therapeutic perspectives. Cancers 12, 3029 (2020).
Francavilla, C. & O’Brien, C. S. Fibroblast growth factor receptor signalling dysregulation and targeting in breast cancer. Open Biol. 12, 210373 (2022).
Mahipal, A., Tella, S. H., Kommalapati, A., Anaya, D. & Kim, R. FGFR2 genomic aberrations: achilles heel in the management of advanced cholangiocarcinoma. Cancer Treat. Rev. 78, 1–7 (2019).
Vogel, A., Segatto, O., Stenzinger, A. & Saborowski, A. FGFR2 inhibition in cholangiocarcinoma. Annu. Rev. Med. 74, 293–306 (2023).
Ilyas, S. I. et al. Cholangiocarcinoma — novel biological insights and therapeutic strategies. Nat. Rev. Clin. Oncol. 20, 470–486 (2023).
Pearson, A. et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 6, 838–851 (2016).
Jogo, T. et al. Circulating tumor DNA analysis detects FGFR2 amplification and concurrent genomic alterations associated with FGFR inhibitor efficacy in advanced gastric cancer. Clin. Cancer Res. 27, 5619–5627 (2021).
Zingg, D. et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature 608, 609–617 (2022).
Wang, H. et al. Advances of fibroblast growth factor/receptor signaling pathway in hepatocellular carcinoma and its pharmacotherapeutic targets. Front. Pharmacol. 12, 650388 (2021).
Gadaleta, R. M. & Moschetta, A. Dark and bright side of targeting fibroblast growth factor receptor 4 in the liver. J. Hepatol. 75, 1440–1451 (2021).
Tao, Z., Cui, Y., Xu, X. & Han, T. FGFR redundancy limits the efficacy of FGFR4-selective inhibitors in hepatocellular carcinoma. Proc. Natl Acad. Sci. USA 119, e2208844119 (2022).
Weiss, J. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2, 62ra93 (2010).
Zhou, Z. et al. Targeting FGFR in non-small cell lung cancer: implications from the landscape of clinically actionable aberrations of FGFR kinases. Cancer Biol. Med. 18, 490–501 (2021).
Pacini, L., Jenks, A. D., Lima, N. C. & Huang, P. H. Targeting the fibroblast growth factor receptor (FGFR) family in lung cancer. Cells 10, 1154 (2021).
Pollock, P. M. et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene 26, 7158–7162 (2007).
Guo, F., Zhang, H., Jia, Z., Cui, M. & Tian, J. Chemoresistance and targeting of growth factors/cytokines signalling pathways: towards the development of effective therapeutic strategy for endometrial cancer. Am. J. Cancer Res. 8, 1317–1331 (2018).
Zhu, D. L., Tuo, X. M., Rong, Y., Zhang, K. & Guo, Y. Fibroblast growth factor receptor signaling as therapeutic targets in female reproductive system cancers. J. Cancer 11, 7264–7275 (2020).
André, F. & Cortés, J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res. Treat. 150, 1–8 (2015).
Papadopoulos, K. P. et al. A phase 1 study of ARQ 087, an oral pan-FGFR inhibitor in patients with advanced solid tumours. Br. J. Cancer 117, 1592–1599 (2017).
Perera, T. P. S. et al. Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol. Cancer Ther. 16, 1010–1020 (2017).
Meric-Bernstam, F. et al. Futibatinib, an irreversible FGFR1-4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: a phase i dose-expansion study. Cancer Discov. 12, 402–415 (2022).
Grünewald, S. et al. Rogaratinib: a potent and selective pan-FGFR inhibitor with broad antitumor activity in FGFR-overexpressing preclinical cancer models. Int. J. Cancer 145, 1346–1357 (2019).
Tyhonas, J. S. et al. Discovery of KIN-3248, an irreversible, next generation FGFR inhibitor for the treatment of advanced tumors harboring FGFR2 and/or FGFR3 gene alterations. J. Med. Chem. 3, 1734–1746 (2024).
Liu, P. C. C. et al. INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS ONE 15, e0231877 (2020).
Guagnano, V. et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2, 1118–1133 (2012).
Gavine, P. R. et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056 (2012).
Nakanishi, Y. et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol. Cancer Ther. 13, 2547–2558 (2014).
Nguyen, M. H. et al. Discovery of orally bioavailable FGFR2/FGFR3 dual inhibitors via structure-guided scaffold repurposing approach. ACS Med. Chem. Lett. 14, 312–318 (2023).
Subbiah, V. et al. RLY-4008, the first highly selective FGFR2 inhibitor with activity across FGFR2 alterations and resistance mutations. Cancer Discov. 13, 2012–2031 (2023).
Ballard, J. A. et al. Preclinical characterization of LOXO-435 (LOX-24350), a potent and highly isoform-selective FGFR3 inhibitor. Mol. Cancer Ther. 20, P141 (2021).
Weiss, A. et al. FGF401, a first-in-class highly selective and potent FGFR4 inhibitor for the treatment of FGF19-driven hepatocellular cancer. Mol. Cancer Ther. 18, 2194–2206 (2019).
Hatlen, M. A. et al. Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discov. 9, 1686–1695 (2019).
Tan, L. et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc. Natl Acad. Sci. USA 111, E4869–E4877 (2014).
FDA. Approval of erdafitinib. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/212018s000lbl.pdf (2019).
FDA. Approval of eutibatinib. https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/214801s000lbl.pdf (2022).
FDA. Approval of infigratinib. https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/214622s000lbl.pdf (2021).
FDA. Approval of pemigatinib. https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213736s002lbl.pdf (2022).
Siefker-Radtke, A. O. et al. Management of fibroblast growth factor inhibitor treatment-emergent adverse events of interest in patients with locally advanced or metastatic urothelial carcinoma. Eur. Urol. Open. Sci. 50, 1–9 (2023).
FDA. FDA approves erdafitinib for locally advanced or metastatic urothelial carcinoma. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-erdafitinib-locally-advanced-or-metastatic-urothelial-carcinoma (2024).
National Comprehensive Cancer Network. NCCN Guidelines. Bladder cancer, version 3.2023, https://www.nccn.org/professionals/physician_gls/pdf/bladder.pdf (2023).
NICE. Inspection of erdafitinib. https://www.nice.org.uk/guidance/indevelopment/gid-ta10937 (2023).
Pant, S. et al. Erdafitinib in patients with advanced solid tumours with FGFR alterations (RAGNAR): an international, single-arm, phase 2 study. Lancet Oncol. 24, 925–935 (2023).
Goyal, L. et al. Primary results of phase 2 FOENIX-CCA2: the irreversible FGFR1-4 inhibitor futibatinib in intrahepatic cholangiocarcinoma (iCCA) with FGFR2 fusions/rearrangements. Cancer Res. 81, CT010 (2021).
Alekseev, O., Ojuok, E. & Cousins, S. Multifocal serous retinopathy with pemigatinib therapy for metastatic colon adenocarcinoma. Int. J. Retin. Vitreous 7, 34 (2021).
Oladipupo, S. S. et al. Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proc. Natl Acad. Sci. USA 111, 13379–13384 (2014).
National Comprehensive Cancer Network. NCCN Guidelines. Biliary tract cancers, version 2.2023, https://www.nccn.org/professionals/physician_gls/pdf/btc.pdf (2023).
European Medicines Agency. Pemazyre (Pemigatinib). 12/09/2023, https://www.ema.europa.eu/en/medicines/human/EPAR/pemazyre (2023).
European Medicines Agency. Lytgobi (Futibatinib). 18/07/2023, https://www.ema.europa.eu/en/medicines/human/EPAR/lytgobi (2023).
National Comprehensive Cancer Network. NCCN Guidelines. Myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions, https://www.nccn.org/professionals/physician_gls/pdf/mlne.pdf (2023).
Loriot, Y. et al. Erdafitinib or chemotherapy in advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 389, 1961–1971 (2023).
Janssen. Press release for erdafitinib. investor.jnj.com, https://www.investor.jnj.com/files/doc_news/2023/09/BALVERSA-Filing-Press-Release_FINAL.pdf (2023).
Catenacci, D. V. T. et al. Phase I escalation and expansion study of bemarituzumab (FPA144) in patients with advanced solid tumors and FGFR2b-selected gastroesophageal adenocarcinoma. J. Clin. Oncol. 38, 2418–2426 (2020).
Wainberg, Z. A. et al. Bemarituzumab in patients with FGFR2b-selected gastric or gastro-oesophageal junction adenocarcinoma (FIGHT): a randomised, double-blind, placebo-controlled, phase 2 study. Lancet Oncol. 23, 1430–1440 (2022).
Wainberg, Z. A. et al. Bemarituzumab as first-line treatment for locally advanced or metastatic gastric/gastroesophageal junction adenocarcinoma: final analysis of the randomized phase 2 FIGHT trial. Gastric Cancer, https://doi.org/10.1007/s10120-024-01466-w (2024).
Schuler, M. et al. Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 20, 1454–1466 (2019).
Voss, M. H. et al. A phase I, open-label, multicenter, dose-escalation study of the oral selective FGFR inhibitor debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clin. Cancer Res. 25, 2699–2707 (2019).
Kommalapati, A. et al. FGFR inhibitors in oncology: insight on the management of toxicities in clinical practice. Cancers 13, 2968 (2021).
Xie, Y. et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 5, 181 (2020).
Han, X. et al. Conditional deletion of Fgfr1 in the proximal and distal tubule identifies distinct roles in phosphate and calcium transport. PLoS ONE 11, e0147845 (2016).
Kim, R. D. et al. First-in-human phase I study of fisogatinib (BLU-554) validates aberrant FGF19 signaling as a driver event in hepatocellular carcinoma. Cancer Discov. 9, 1696–1707 (2019).
Chan, S. L. et al. A first-in-human phase 1/2 study of FGF401 and combination of FGF401 with spartalizumab in patients with hepatocellular carcinoma or biomarker-selected solid tumors. J. Exp. Clin. Cancer Res. 41, 189 (2022).
Cowell, J. K. et al. Mutation in the FGFR1 tyrosine kinase domain or inactivation of PTEN is associated with acquired resistance to FGFR inhibitors in FGFR1-driven leukemia/lymphomas. Int. J. Cancer 141, 1822–1829 (2017).
Pal, S. K. et al. Efficacy of BGJ398, a fibroblast growth factor receptor 1–3 inhibitor, in patients with previously treated advanced urothelial carcinoma with FGFR3 alterations. Cancer Discov. 8, 812–821 (2018).
Goyal, L. et al. TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov. 9, 1064–1079 (2019).
Varghese, A. M. et al. Noninvasive detection of polyclonal acquired resistance to FGFR inhibition in patients with cholangiocarcinoma harboring FGFR2 alterations. JCO Precis. Oncol. 5, 44–50 (2021).
Silverman, I. M. et al. Clinicogenomic analysis of FGFR2-rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov. 11, 326–339 (2021).
Jiang, K. et al. GZD824 overcomes FGFR1-V561F/M mutant resistance in vitro and in vivo. Cancer Med. 10, 4874–4884 (2021).
Valery, M. et al. Targetable molecular alterations in the treatment of biliary tract cancers: an overview of the available treatments. Cancers 15, 4446 (2023).
Facchinetti, F. et al. Resistance to selective FGFR inhibitors in FGFR-driven urothelial cancer. Cancer Discov. 13, 1998–2011 (2023).
Guercio, B. J. et al. Clinical and genomic landscape of FGFR3-altered urothelial carcinoma and treatment outcomes with erdafitinib: a real-world experience. Clin. Cancer Res. 29, 4586–4595 (2023).
Wu, Q. et al. Landscape of clinical resistance mechanisms to FGFR inhibitors in FGFR2-altered cholangiocarcinoma. Clin. Cancer Res. 30, 198–208 (2024).
Mohammadi, M., Schlessinger, J. & Hubbard, S. R. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 86, 577–587 (1996).
Rizzo, A., Ricci, A. D. & Brandi, G. Futibatinib, an investigational agent for the treatment of intrahepatic cholangiocarcinoma: evidence to date and future perspectives. Expert Opin. Investig. Drugs 30, 317–324 (2021).
Murugesan, K. et al. Pan-tumor landscape of fibroblast growth factor receptor 1–4 genomic alterations. ESMO Open 7, 100641 (2022).
Schram, A. M. et al. Clinical activity of lirafugratinib (RLY-4008), a highly selective FGFR2 inhibitor, in patients with advanced FGFR2-altered solid tumors: the ReFocus study. Mol. Cancer Ther. 22, IA006 (2023).
Iyer, G. et al. A first-in-human phase 1 study of LOXO-435, a potent, highly isoform-selective FGFR3 inhibitor in advanced solid tumors with FGFR3 alterations (trial in progress). Cancer Res. 83, CT119 (2023).
Herrera-Abreu, M. T. et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3-mutant cancer. Cancer Discov. 3, 1058–1071 (2013).
Wu, Q. et al. EGFR inhibition potentiates FGFR inhibitor therapy and overcomes resistance in FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 12, 1378–1395 (2022).
Siefker-Radtke, A. O. et al. Analysis of circulating tumor DNA (ctDNA) from the phase II BLC2001 trial of erdafitinib in locally advanced or metastatic urothelial carcinoma (mUC) to identify markers of intrinsic resistance to fibroblast growth factor receptor (FGFR)-targeted therapy. Ann. Oncol. 31, S584 (2020).
Cleary, J. M. et al. FGFR2 extracellular domain in-frame deletions are therapeutically targetable genomic alterations that function as oncogenic drivers in cholangiocarcinoma. Cancer Discov. 11, 2488–2505 (2021).
Malchers, F. et al. Mechanisms of primary drug resistance in FGFR1-amplified lung cancer. Clin. Cancer Res. 23, 5527–5536 (2017).
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 15, 139–150 (2018).
Donehower, L. A. et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 28, 1370–1384 (2019).
Baslan, T. et al. Ordered and deterministic cancer genome evolution after p53 loss. Nature 608, 795–802 (2022).
Wang, X. Y. et al. Driver mutations of intrahepatic cholangiocarcinoma shape clinically relevant genomic clusters with distinct molecular features and therapeutic vulnerabilities. Theranostics 12, 260–276 (2022).
Tavolari, S. & Brandi, G. Mutational landscape of cholangiocarcinoma according to different etiologies: a review. Cells 12, 1216 (2023).
Subbiah, V. et al. FIGHT-101, a first-in-human study of potent and selective FGFR 1-3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann. Oncol. 33, 522–533 (2022).
Kamoun, A. et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur. Urol. 77, 420–433 (2020).
Mata, D. A. et al. Genetic and epigenetic landscape of IDH-wildtype glioblastomas with FGFR3–TACC3 fusions. Acta Neuropathol. Commun. 8, 186 (2020).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Chirnomas, D., Hornberger, K. R. & Crews, C. M. Protein degraders enter the clinic — a new approach to cancer therapy. Nat. Rev. Clin. Oncol. 20, 265–278 (2023).
He, M. et al. PROTACs: great opportunities for academia and industry (an update from 2020 to 2021). Signal Transduct. Target. Ther. 7, 181 (2022).
Ma, L. et al. Discovery of a selective and orally bioavailable FGFR2 degrader for treating gastric cancer. J. Med. Chem. 66, 7438–7453 (2023).
Du, G. et al. Discovery of a potent degrader for fibroblast growth factor receptor 1/2. Angew. Chem. Int. Ed. Engl. 60, 15905–15911 (2021).
Larson, R. C. & Maus, M. V. Recent advances and discoveries in the mechanisms and functions of CAR T cells. Nat. Rev. Cancer 21, 145–161 (2021).
Maalej, K. M. et al. CAR-cell therapy in the era of solid tumor treatment: current challenges and emerging therapeutic advances. Mol. Cancer 22, 20 (2023).
Sullivan, P. M. et al. FGFR4-targeted chimeric antigen receptors combined with anti-myeloid polypharmacy effectively treat orthotopic rhabdomyosarcoma. Mol. Cancer Ther. 21, 1608–1621 (2022).
Tian, M. et al. Preclinical development of a chimeric antigen receptor T cell therapy targeting FGFR4 in rhabdomyosarcoma. Cell Rep. Med. 4, 101212 (2023).
Sun, H. D. et al. Monoclonal antibody antagonists of hypothalamic FGFR1 cause potent but reversible hypophagia and weight loss in rodents and monkeys. Am. J. Physiol. Endocrinol. Metab. 292, E964–E976 (2007).
Sommer, A. et al. Preclinical efficacy of the auristatin-based antibody-drug conjugate BAY 1187982 for the treatment of FGFR2-positive solid tumors. Cancer Res. 76, 6331–6339 (2016).
Kamath, A. V. et al. Preclinical pharmacokinetics of MFGR1877A, a human monoclonal antibody to FGFR3, and prediction of its efficacious clinical dose for the treatment of t(4;14)-positive multiple myeloma. Cancer Chemother. Pharmacol. 69, 1071–1078 (2012).
Bartz, R. et al. Preclinical development of U3-1784, a novel FGFR4 antibody against cancer, and avoidance of its on-target toxicity. Mol. Cancer Ther. 18, 1832–1843 (2019).
Drago, J. Z., Modi, S. & Chandarlapaty, S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol. 18, 327–344 (2021).
Fu, Z. et al. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Signal Transduct. Target. Ther. 7, 93 (2022).
Kollmannsberger, C. et al. A phase 1 study of LY3076226, a fibroblast growth factor receptor 3 (FGFR3) antibody-drug conjugate, in patients with advanced or metastatic cancer. Invest. New Drugs 39, 1613–1623 (2021).
Kim, S. B. et al. First-in-human phase I study of aprutumab ixadotin, a fibroblast growth factor receptor 2 antibody-drug conjugate (BAY 1187982) in patients with advanced cancer. Target. Oncol. 14, 591–601 (2019).
Wickstroem, K. et al. Preclinical combination studies of an FGFR2 targeted thorium-227 conjugate and the ATR inhibitor BAY 1895344. Int. J. Radiat. Oncol. Biol. Phys. 105, 410–422 (2019).
Nasr, D. et al. Radioimmunoconjugates in the age of modern immuno-oncology. Life Sci. 310, 121126 (2022).
Harding, T. C. et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci. Transl. Med. 5, 178ra39 (2013).
Tolcher, A. W. et al. A phase I, first in human study of FP-1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Ann. Oncol. 27, 526–532 (2016).
Morgensztern, D. et al. An open-label phase IB study to evaluate GSK3052230 in combination with paclitaxel and carboplatin, or docetaxel, in FGFR1-amplified non-small cell lung cancer. Lung Cancer 136, 74–79 (2019).
van Brummelen, E. M. J. et al. A phase Ib study of GSK3052230, an FGF ligand trap in combination with pemetrexed and cisplatin in patients with malignant pleural mesothelioma. Invest. New Drugs 38, 457–467 (2020).
Gonçalves, D. et al. In vitro and in vivo characterization of recifercept, a soluble fibroblast growth factor receptor 3, as treatment for achondroplasia. PLoS ONE 15, e0244368 (2020).
Ishiwata, T. Role of fibroblast growth factor receptor-2 splicing in normal and cancer cells. Front. Biosci. 23, 626–639 (2018).
Epstein, R. J., Tian, L. J. & Gu, Y. F. 2b or not 2b: how opposing FGF receptor splice variants are blocking progress in precision oncology. J. Oncol. 2021, 9955456 (2021).
Taylor, J. G. VI et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Invest. 119, 3395–3407 (2009).
Sartor, O. et al. Lutetium-177-PSMA-617 for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 385, 1091–1103 (2021).
Yao, J. et al. Synthesis and evaluation of novel 99mTc-labeled FGFR2-targeting peptides. Mol. Pharm. 21, 895–903 (2024).
Lin, C. C. et al. Receptor tyrosine kinases regulate signal transduction through a liquid-liquid phase separated state. Mol. Cell 82, 1089–1106 (2022).
Hart, K. C., Robertson, S. C. & Donoghue, D. J. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3-kinase activation. Mol. Biol. Cell 12, 931–942 (2001).
Szybowska, P., Kostas, M., Wesche, J., Wiedlocha, A. & Haugsten, E. M. Cancer mutations in FGFR2 prevent a negative feedback loop mediated by the ERK1/2 pathway. Cells 8, 518 (2019).
Turner, N. et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 70, 2085–2094 (2010).
Razavi, P. et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438 (2018).
Sircoulomb, F. et al. ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol. Med. 3, 153–166 (2011).
Rutkovsky, A. C. et al. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer 19, 491 (2019).
Yuan, G. et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 590, 504–508 (2021).
Condorelli, R. et al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 30, 365–373 (2019).
Yao, Q. et al. Epigenetic alterations in keratinocyte carcinoma. J. Invest. Dermatol. 141, 1207–1218 (2021).
DeSalvo, J. et al. ETV4 and ETV5 drive synovial sarcoma through cell cycle and DUX4 embryonic pathway control. J. Clin. Invest. 131, e141908 (2021).
Lötsch, D. et al. Targeting fibroblast growth factor receptors to combat aggressive ependymoma. Acta Neuropathol. 142, 339–360 (2021).
Mouron, S. et al. FGFR1 amplification or overexpression and hormonal resistance in luminal breast cancer: rationale for a triple blockade of ER, CDK4/6, and FGFR1. Breast Cancer Res. 23, 21 (2021).
Formisano, L. et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 10, 1373 (2019).
Coombes, R. C. et al. Results of the phase IIa RADICAL trial of the FGFR inhibitor AZD4547 in endocrine resistant breast cancer. Nat. Commun. 13, 3246 (2022).
Mayer, I. A. et al. A phase Ib trial of fulvestrant + CDK4/6 inhibitor (CDK4/6i) palbociclib + pan-FGFR tyrosine kinase inhibitor (TKI) erdafitinib in FGFR-amplified/ ER+/ HER2- negative metastatic breast cancer (MBC). Cancer Res. 81, PD1–PD03 (2021).
Giacomini, A. et al. The FGF/FGFR system in the physiopathology of the prostate gland. Physiol. Rev. 101, 569–610 (2021).
Bluemn, E. G. et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489 (2017).
Cheng, C. et al. Gremlin1 is a therapeutically targetable FGFR1 ligand that regulates lineage plasticity and castration resistance in prostate cancer. Nat. Cancer 3, 565–580 (2022).
Chan, J. M. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 377, 1180–1191 (2022).
Chiodelli, P. et al. FGFR blockade by pemigatinib treats naïve and castration resistant prostate cancer. Cancer Lett. 526, 217–224 (2022).
Kraehenbuehl, L., Weng, C. H., Eghbali, S., Wolchok, J. D. & Merghoub, T. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat. Rev. Clin. Oncol. 19, 37–50 (2022).
Cristescu, R. et al. Transcriptomic determinants of response to pembrolizumab monotherapy across solid tumor types. Clin. Cancer Res. 28, 1680–1689 (2022).
Petroni, G., Buqué, A., Coussens, L. M. & Galluzzi, L. Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat. Rev. Drug. Discov. 21, 440–462 (2022).
Jing, W. et al. FGFR3 destabilizes PD-L1 via NEDD4 to control T-cell-mediated bladder cancer immune surveillance. Cancer Res. 82, 114–129 (2022).
Ouyang, Y. et al. FGFR3 alterations in bladder cancer stimulate serine synthesis to induce immune-inert macrophages that suppress T-cell recruitment and activation. Cancer Res. 83, 4030–4046 (2023).
Katoh, M. & Katoh, M. WNT signaling and cancer stemness. Essays Biochem. 66, 319–331 (2022).
Huinen, Z. R., Huijbers, E. J. M., van Beijnum, J. R., Nowak-Sliwinska, P. & Griffioen, A. W. Anti-angiogenic agents — overcoming tumour endothelial cell anergy and improving immunotherapy outcomes. Nat. Rev. Clin. Oncol. 18, 527–540 (2021).
Davidson, S. et al. Fibroblasts as immune regulators in infection, inflammation and cancer. Nat. Rev. Immunol. 21, 704–717 (2021).
Ruan, R. et al. Unleashing the potential of combining FGFR inhibitor and immune checkpoint blockade for FGF/FGFR signaling in tumor microenvironment. Mol. Cancer 22, 60 (2023).
Siefker-Radtke, A. O. et al. Erdafitinib versus pembrolizumab in pretreated patients with advanced or metastatic urothelial cancer with select FGFR alterations: cohort 2 of the randomized phase III THOR trial. Ann. Oncol. 35, 107–117 (2024).
Song, Y. et al. Fibroblast growth factor receptor 3 mutation attenuates response to immune checkpoint blockade in metastatic urothelial carcinoma by driving immunosuppressive microenvironment. J. Immunother. Cancer 11, e006643 (2023).
Okato, A. et al. FGFR inhibition augments anti-PD-1 efficacy in murine FGFR3-mutant bladder cancer by abrogating immunosuppression. J. Clin. Invest. 134, e169241 (2024).
Rose, T. L. et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br. J. Cancer 125, 1251–1260 (2021).
Siefker-Radtke, A. O. et al. Erdafitinib (ERDA) vs ERDA plus cetrelimab (ERDA+CET) for patients (pts) with metastatic urothelial carcinoma (mUC) and fibroblast growth factor receptor alterations (FGFRa): final results from the phase 2 Norse study. J. Clin. Oncol. 41, 4504 (2023).
Koshkin, V. S. et al. Futibatinib plus pembrolizumab in patients (pts) with advanced or metastatic urothelial carcinoma (mUC): preliminary safety results from a phase 2 study. J. Clin. Oncol. 40, 501 (2022).
Rosenberg, J. E. et al. Safety and efficacy of rogaratinib in combination with atezolizumab in cisplatin-ineligible patients (pts) with locally advanced or metastatic urothelial cancer (UC) and FGFR mRNA overexpression in the phase Ib/II FORT-2 study. J. Clin. Oncol. 39, 4521 (2021).
Powles, T. B. et al. Erdafitinib (ERDA) or ERDA plus cetrelimab (CET) for patients with metastatic or locally advanced urothelial carcinoma (mUC) and fibroblast growth factor receptor alterations (FGFRa): first phase (Ph) II results from the NORSE study. Ann. Oncol. 32, S1303 (2021).
Deshmukh, A. P. et al. Identification of EMT signaling cross-talk and gene regulatory networks by single-cell RNA sequencing. Proc. Natl Acad. Sci. USA 118, e2102050118 (2021).
Oh, S. C. et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat. Commun. 9, 1777 (2018).
Dong, D. et al. FZD5 prevents epithelial-mesenchymal transition in gastric cancer. Cell Commun. Signal. 19, 21 (2021).
Ho, S. W. T. et al. Regulatory enhancer profiling of mesenchymal-type gastric cancer reveals subtype-specific epigenomic landscapes and targetable vulnerabilities. Gut 72, 226–241 (2023).
Warzecha, C. C., Sato, T. K., Nabet, B., Hogenesch, J. B. & Carstens, R. P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 33, 591–601 (2009).
Zhao, Y. et al. Downregulated ESRP1/2 promotes lung metastasis of bladder carcinoma through altering FGFR2 splicing and macrophage polarization. Front. Immunol. 14, 1161273 (2023).
Ashok, C. et al. E2F1 and epigenetic modifiers orchestrate breast cancer progression by regulating oxygen-dependent ESRP1 expression. Oncogenesis 10, 58 (2021).
Groulx, J. F., Boudjadi, S. & Beaulieu, J. F. MYC regulates α6 integrin subunit expression and splicing under its pro-proliferative ITGA6A form in colorectal cancer cells. Cancers 10, 42 (2018).
Gemmill, R. M. et al. ZEB1-responsive genes in non-small cell lung cancer. Cancer Lett. 300, 66–78 (2011).
Reinke, L. M., Xu, Y. & Cheng, C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J. Biol. Chem. 287, 36435–36442 (2012).
Brown, W. S., Tan, L., Smith, A., Gray, N. S. & Wendt, M. K. Covalent targeting of fibroblast growth factor receptor inhibits metastatic breast cancer. Mol. Cancer Ther. 15, 2096–2106 (2016).
Sengal, A. T. et al. Spatial expression of the FGFR2b splice isoform and its prognostic significance in endometrioid endometrial carcinoma. J. Pathol. Clin. Res. 8, 521–537 (2022).
Yashiro, M. et al. Clinical difference between fibroblast growth factor receptor 2 subclass, type IIIb and type IIIc, in gastric cancer. Sci. Rep. 11, 4698 (2021).
Minashi, K. et al. Cancer-related FGFR2 overexpression and gene amplification in Japanese patients with gastric cancer. Jpn J. Clin. Oncol. 51, 1523–1533 (2021).
Neumann, O. et al. Genomic architecture of FGFR2 fusions in cholangiocarcinoma and its implication for molecular testing. Br. J. Cancer 127, 1540–1549 (2022).
Katoh, M. The integration of genomics testing and functional proteomics in the era of personalized medicine. Expert Rev. Proteom. 14, 1055–1058 (2017).
Halldorsson, B. V. et al. The sequences of 150,119 genomes in the UK Biobank. Nature 607, 732–740 (2022).
Martínez-Jiménez, F. et al. Pan-cancer whole-genome comparison of primary and metastatic solid tumours. Nature 618, 333–341 (2023).
Sternberg, C. N. et al. FORT-1: phase II/III study of rogaratinib versus chemotherapy in patients with locally advanced or metastatic urothelial carcinoma selected based on FGFR1/3 mRNA expression. J. Clin. Oncol. 41, 629–639 (2022).
Velmahos, C. S., Badgeley, M. & Lo, Y. C. Using deep learning to identify bladder cancers with FGFR-activating mutations from histology images. Cancer Med. 10, 4805–4813 (2021).
Loeffler, C. M. L. et al. Artificial intelligence-based detection of FGFR3 mutational status directly from routine histology in bladder cancer: a possible preselection for molecular testing? Eur. Urol. Focus 8, 472–479 (2022).
Mohammadi, M. et al. Structures of the tyrosine-kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 (1997).
Acknowledgements
This work was partly supported by grants-in-aid from Masaru Katoh’s Fund for Knowledge-Base and Global Network Projects (Masuko Katoh and Masaru Katoh) and Fondazione Donato-Venturi (G.B.).
Author information
Authors and Affiliations
Contributions
Masuko Katoh and Masaru Katoh researched data for the manuscript, made substantial contributions to discussions of content, and wrote the manuscript. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
Y.L. has acted as a consultant and/or adviser for Astellas, AstraZeneca, Bristol Myers Squibb, Gilead, Janssen, Merck KGaA, Pfizer, Roche, Seattle Genetics and Taiho and has received research grants from Celsius, Janssen and Sanofi. G.B. has acted as an adviser for Incyte, Lilly and Taiho and has received research grants from Incyte and Ipsen. Z.A.W. has been a consultant for Amgen, Arcus, Astellas, AstraZeneca, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Gilead, Ipsen, Merck, Novartis and Seagen, has received research grants from Arcus, Bristol Myers Squibb and Plexxikon, and has served on the data and safety monitoring boards of Compass, Daiichi Sankyo, Mirati and Pfizer.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks J. Garcia-Donas who co-reviewed with T. Grazioso, Jacek Otlewski who co-reviewed with Malgorzata Zakrzewska, Sumanta Pal, Roberto Ronca and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
cBioportal: https://www.cbioportal.org
ClinicalTrials.gov: https://clinicaltrials.gov
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Katoh, M., Loriot, Y., Brandi, G. et al. FGFR-targeted therapeutics: clinical activity, mechanisms of resistance and new directions. Nat Rev Clin Oncol 21, 312–329 (2024). https://doi.org/10.1038/s41571-024-00869-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-024-00869-z