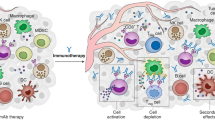
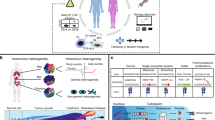
Abstract
Despite the notable success of chimeric antigen receptor (CAR) T cell therapies in the treatment of certain haematological malignancies, challenges remain in optimizing CAR designs and cell products, improving response rates, extending the durability of remissions, reducing toxicity and broadening the utility of this therapeutic modality to other cancer types. Data from multidimensional omics analyses, including genomics, epigenomics, transcriptomics, T cell receptor-repertoire profiling, proteomics, metabolomics and/or microbiomics, provide unique opportunities to dissect the complex and dynamic multifactorial phenotypes, processes and responses of CAR T cells as well as to discover novel tumour targets and pathways of resistance. In this Review, we summarize the multidimensional cellular and molecular profiling technologies that have been used to advance our mechanistic understanding of CAR T cell therapies. In addition, we discuss current applications and potential strategies leveraging multi-omics data to identify optimal target antigens and other molecular features that could be exploited to enhance the antitumour activity and minimize the toxicity of CAR T cell therapy. Indeed, fully utilizing multi-omics data will provide new insights into the biology of CAR T cell therapy, further accelerate the development of products with improved efficacy and safety profiles, and enable clinicians to better predict and monitor patient responses.
Key points
-
Multidimensional omics data, encompassing genomics, epigenomics, transcriptomics, T cell receptor-repertoire profiling, proteomics, metabolomics and microbiomics, have been exploited to advance our mechanistic understanding of chimeric antigen receptor (CAR) T cell therapy.
-
Utilization of multi-omics data derived from both tumour and non-malignant tissues at the bulk and/or single-cell levels is a powerful approach to identifying optimal targets for highly efficacious and safe CAR T cell therapy.
-
Integration of bulk and/or single-cell multidimensional omics data has been applied to investigate key determinants of CAR T cell persistence and antitumour efficacy, including T cell states and phenotypes, tumour cell characteristics, the tumour microenvironment and the microbiome.
-
Leveraging multi-omics data promises to elucidate the mechanisms underlying CAR T cell-related toxicities, including cytokine-release syndrome, immune effector cell-associated neurotoxicity syndrome and on-target, off-tumour toxicities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020).
Maude, S. L. et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014).
Schuster, S. J. et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N. Engl. J. Med. 377, 2545–2554 (2017).
Wang, M. et al. KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 382, 1331–1342 (2020).
Fowler, N. H. et al. Tisagenlecleucel in adult relapsed or refractory follicular lymphoma: the phase 2 ELARA trial. Nat. Med. 28, 325–332 (2022).
Berdeja, J. G. et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 398, 314–324 (2021).
Brown, C. E. et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N. Engl. J. Med. 375, 2561–2569 (2016).
Beatty, G. L. et al. Activity of mesothelin-specific chimeric antigen receptor T cells against pancreatic carcinoma metastases in a phase 1 trial. Gastroenterology 155, 29–32 (2018).
Ahmed, N. et al. Human epidermal growth factor receptor 2 (HER2)-specific chimeric antigen receptor-modified T cells for the immunotherapy of HER2-positive sarcoma. J. Clin. Oncol. 33, 1688–1696 (2015).
Qi, C. et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat. Med. 28, 1189–1198 (2022).
Narayan, V. et al. PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: a phase 1 trial. Nat. Med. 28, 724–734 (2022).
Irving, M., Zoete, V., Bassani-Sternberg, M. & Coukos, G. A roadmap for driving CAR T cells toward the oncogenic immunopeptidome. Cancer Cell 40, 20–22 (2022).
Leko, V. & Rosenberg, S. A. Identifying and targeting human tumor antigens for T cell-based immunotherapy of solid tumors. Cancer Cell 38, 454–472 (2020).
Delgoffe, G. M. et al. The role of exhaustion in CAR T cell therapy. Cancer Cell 39, 885–888 (2021).
Deng, Q. et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020).
Hou, A. J., Chen, L. C. & Chen, Y. Y. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov. 20, 531–550 (2021).
Milone, M. C. et al. Engineering-enhanced CAR T cells for improved cancer therapy. Nat. Cancer 2, 780–793 (2021).
Neelapu, S. S. et al. Chimeric antigen receptor T-cell therapy-assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15, 47–62 (2018).
Santomasso, B. D. et al. Management of immune-related adverse events in patients treated with chimeric antigen receptor T-cell therapy: ASCO guideline. J. Clin. Oncol. 39, 3978–3992 (2021).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 83 (2017).
Deng, Y., Finck, A. & Fan, R. Single-cell omics analyses enabled by microchip technologies. Annu. Rev. Biomed. Eng. 21, 365–393 (2019).
Cantini, L. et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat. Commun. 12, 124 (2021).
Jiang, P. et al. Big data in basic and translational cancer research. Nat. Rev. Cancer https://doi.org/10.1038/s41568-022-00502-0 (2022).
Haradhvala, N. J. et al. Distinct cellular dynamics associated with response to CAR-T therapy for refractory B cell lymphoma. Nat. Med. 28, 1848–1859 (2022).
Scholler, N. et al. Tumor immune contexture is a determinant of anti-CD19 CAR T cell efficacy in large B cell lymphoma. Nat. Med. https://doi.org/10.1038/s41591-022-01916-x (2022).
Good, Z. et al. Post-infusion CAR Treg cells identify patients resistant to CD19-CAR therapy. Nat. Med. 28, 1860–1871 (2022).
Young, R. M., Engel, N. W., Uslu, U., Wellhausen, N. & June, C. H. Next-generation CAR T-cell therapies. Cancer Discov. 12, 1625–1633 (2022).
Chen, G. M. et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 11, 2186–2199 (2021).
Chen, J. et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019).
Fraietta, J. A. et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018).
Xhangolli, I. et al. Single-cell analysis of CAR-T cell activation reveals a mixed TH1/TH2 response independent of differentiation. Genomics Proteomics Bioinformatics 17, 129–139 (2019).
Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019).
Garcia-Prieto, C. A. et al. Epigenetic profiling and response to CD19 chimeric antigen receptor T-cell therapy in B-cell malignancies. J. Natl Cancer Inst. 114, 436–445 (2022).
Ye, L. et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab. 34, 595–614.e14 (2022).
Ma, W. et al. Targeting PAK4 to reprogram the vascular microenvironment and improve CAR-T immunotherapy for glioblastoma. Nat. Cancer 2, 83–97 (2021).
Norelli, M. et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 24, 739–748 (2018).
Giavridis, T. et al. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 24, 731–738 (2018).
MacKay, M. et al. The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat. Biotechnol. 38, 233–244 (2020).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Yamamoto, T. N., Kishton, R. J. & Restifo, N. P. Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat. Med. 25, 1488–1499 (2019).
Hwang, M. S. et al. Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens. Nat. Commun. 12, 5271 (2021).
Choe, J. H. et al. SynNotch-CAR T cells overcome challenges of specificity, heterogeneity, and persistence in treating glioblastoma. Sci. Transl. Med. 13, eabe7378 (2021).
O’Rourke, D. M. et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017).
Shouval, R. et al. Impact of TP53 genomic alterations in large B-cell lymphoma treated with CD19-chimeric antigen receptor T-cell therapy. J. Clin. Oncol. 40, 369–381 (2022).
Cherng, H.-J. J. et al. Risk assessment with low-pass whole-genome sequencing of cell-free DNA before CD19 CAR T-cell therapy for large B-cell lymphoma. Blood 140, 504–515 (2022).
Jain, M. D. et al. Whole-genome sequencing reveals complex genomic features underlying anti-CD19 CAR T-cell treatment failures in lymphoma. Blood 140, 491–503 (2022).
Sworder, B. J. et al. Determinants of resistance to engineered T cell therapies targeting CD19 in large B cell lymphomas. Cancer Cell https://doi.org/10.1016/j.ccell.2022.12.005 (2022).
Zhou, Y. et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491 (2014).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Prim. https://doi.org/10.1038/s43586-021-00093-4 (2022).
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018).
Wei, J. et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476 (2019).
Wang, D. et al. CRISPR screening of CAR T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov. 11, 1192–1211 (2021).
Dufva, O. et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood 135, 597–609 (2020).
Gurusamy, D. et al. Multi-phenotype CRISPR-Cas9 screen identifies p38 kinase as a target for adoptive immunotherapies. Cancer Cell 37, 818–833.e9 (2020).
Singh, N. et al. Impaired death receptor signaling in leukemia causes antigen-independent resistance by inducing CAR T-cell dysfunction. Cancer Discov. 10, 552–567 (2020).
Yan, X. et al. CD58 loss in tumor cells confers functional impairment of CAR T cells. Blood Adv. 6, 5844–5856 (2022).
Roth, T. L. et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell 181, 728–744.e21 (2020).
Zebley, C. C., Gottschalk, S. & Youngblood, B. Rewriting history: epigenetic reprogramming of CD8+ T cell differentiation to enhance immunotherapy. Trends Immunol. 41, 665–675 (2020).
Henning, A. N., Roychoudhuri, R. & Restifo, N. P. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18, 340–356 (2018).
Akbari, B. et al. Epigenetic strategies to boost CAR T cell therapy. Mol. Ther. 29, 2640–2659 (2021).
Chan, J. D. et al. Cellular networks controlling T cell persistence in adoptive cell therapy. Nat. Rev. Immunol. 21, 769–784 (2021).
Fraietta, J. A. et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018).
Weber, E. W. et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, eaba1786 (2021).
Guo, A. et al. cBAF complex components and MYC cooperate early in CD8+ T cell fate. Nature 607, 135–141 (2022).
Stricker, S. H., Köferle, A. & Beck, S. From profiles to function in epigenomics. Nat. Rev. Genet. 18, 51–66 (2016).
Wang, Y. et al. Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nat. Commun. 12, 409 (2021).
Wei, Y. et al. Targeting CDK7 suppresses super enhancer-linked inflammatory genes and alleviates CAR T cell-induced cytokine release syndrome. Mol. Cancer 20, 5 (2021).
Gennert, D. G. et al. Dynamic chromatin regulatory landscape of human CAR T cell exhaustion. Proc. Natl Acad. Sci. USA 118, e2104758118 (2021).
Ding, Z.-C. et al. Persistent STAT5 activation reprograms the epigenetic landscape in CD4+ T cells to drive polyfunctionality and antitumor immunity. Sci. Immunol. 5, eaba5962 (2020).
Preissl, S., Gaulton, K. J. & Ren, B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00509-1 (2022).
Jiang, P. et al. Single-cell ATAC-seq maps the comprehensive and dynamic chromatin accessibility landscape of CAR-T cell dysfunction. Leukemia https://doi.org/10.1038/s41375-022-01676-0 (2022).
Bagaev, A. et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 39, 845–865.e7 (2021).
Hong, M. et al. RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 13, 166 (2020).
Xiang, Y., Ye, Y., Zhang, Z. & Han, L. Maximizing the utility of cancer transcriptomic data. Trends Cancer 4, 823–837 (2018).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Good, C. R. et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100.e26 (2021).
Michie, J. et al. Antagonism of IAPs enhances CAR T-cell efficacy. Cancer Immunol. Res. 7, 183–192 (2019).
Giuffrida, L. et al. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances CAR T cell efficacy. Nat. Commun. 12, 3236 (2021).
Papalexi, E. et al. Single-cell metabolic profiling of human cytotoxic T cells. Nature 12, 175–188 (2021).
Mineo, M. et al. Tumor interferon signaling is regulated by a lncRNA INCR1 transcribed from the PD-L1 locus. Mol. Cell 78, 1207–1223.e8 (2020).
Sotillo, E. et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015).
Pan, Y. et al. RNA dysregulation: an expanding source of cancer immunotherapy targets. Trends Pharmacol. Sci. 42, 268–282 (2021).
Li, N. et al. CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice. Cell Rep. Med. 2, 100297 (2021).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Armingol, E., Officer, A., Harismendy, O. & Lewis, N. E. Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet. 22, 71–88 (2021).
Suvà, M. L. & Tirosh, I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell 75, 7–12 (2019).
Boroughs, A. C. et al. A distinct transcriptional program in human CAR T cells bearing the 4-1BB signaling domain revealed by scRNA-seq. Mol. Ther. 28, 2577–2592 (2020).
Li, X. X. et al. Single-cell transcriptomic analysis reveals BCMA CAR-T cell dynamics in a patient with refractory primary plasma cell leukemia. Mol. Ther. 29, 645–657 (2021).
Parker, K. R. et al. Single-cell analyses identify brain mural cells expressing CD19 as potential off-tumor targets for CAR-T immunotherapies. Cell 183, 126–142.e17 (2020).
Jing, Y. et al. Expression of chimeric antigen receptor therapy targets detected by single-cell sequencing of normal cells may contribute to off-tumor toxicity. Cancer Cell 39, 1558–1559 (2021).
Boulch, M. et al. A cross-talk between CAR T cell subsets and the tumor microenvironment is essential for sustained cytotoxic activity. Sci. Immunol. 6, eabd4344 (2021).
Dhodapkar, K. M. et al. Changes in bone marrow tumor and immune cells correlate with durability of remissions following BCMA CAR T therapy in myeloma. Blood Cancer Discov. https://doi.org/10.1158/2643-3230.BCD-22-0018 (2022).
Gohil, S. H., Iorgulescu, J. B., Braun, D. A., Keskin, D. B. & Livak, K. J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat. Rev. Clin. Oncol. 18, 244–256 (2021).
Bradley, P. & Thomas, P. G. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol. 37, 547–570 (2019).
Pai, J. A. & Satpathy, A. T. High-throughput and single-cell T cell receptor sequencing technologies. Nat. Methods 18, 881–892 (2021).
Sheih, A. et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020).
Melenhorst, J. J. et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 602, 503–509 (2022).
Mani, D. R. et al. Cancer proteogenomics: current impact and future prospects. Nat. Rev. Cancer 22, 298–313 (2022).
Enblad, G. et al. A phase I/IIa trial using CD19-targeted third-generation CAR T cells for lymphoma and leukemia. Clin. Cancer Res. 24, 6185–6194 (2018).
Hu, Z. et al. The Cancer Surfaceome Atlas integrates genomic, functional and drug response data to identify actionable targets. Nat. Cancer 2, 1406–1422 (2021).
Chong, C., Coukos, G. & Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 40, 175–188 (2022).
Humphrey, S. J., Azimifar, S. B. & Mann, M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 (2015).
Nix, M. A. et al. Surface proteomics reveals CD72 as a target for in vitro–evolved nanobody-based CAR-T cells in KMT2A/MLL1-rearranged B-ALL. Cancer Discov. 11, 2032–2049 (2021).
Perna, F. et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 32, 506–519.e5 (2017).
Lee, J. K. et al. Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc. Natl Acad. Sci. USA 115, E4473–E4482 (2018).
Marcu, A. et al. HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer 9, e002071 (2021).
Salter, A. I. et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci. Signal. 11, eaat6753 (2018).
Ramello, M. C. et al. An immunoproteomic approach to characterize the CAR interactome and signalosome. Sci. Signal. 12, eaap9777 (2019).
Wu, W. et al. Multiple signaling roles of CD3ε and its application in CAR-T cell therapy. Cell 182, 855–871.e23 (2020).
Lombard-Banek, C. & Schiel, J. E. Mass spectrometry advances and perspectives for the characterization of emerging adoptive cell therapies. Molecules 25, 1396 (2020).
Saeys, Y., Van Gassen, S. & Lambrecht, B. N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 16, 449–462 (2016).
Bonilla, D. L., Reinin, G. & Chua, E. Full spectrum flow cytometry as a powerful technology for cancer immunotherapy research. Front. Mol. Biosci. 7, 612801 (2020).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Chong, E. A. et al. Pembrolizumab for B-cell lymphomas relapsing after or refractory to CD19-directed CAR T-cell therapy. Blood 139, 1026–1038 (2022).
Goldberg, L. et al. Single-cell analysis by mass cytometry reveals CD19 CAR T cell spatiotemporal plasticity in patients. Oncoimmunology 11, 2040772 (2022).
Fisher, J. et al. Engineering γδT cells limits tonic signaling associated with chimeric antigen receptors. Sci. Signal. 12, eaax1872 (2019).
Rossi, J. et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 132, 804–814 (2018).
Bai, Z., Su, G. & Fan, R. Single-cell analysis technologies for immuno-oncology research: from mechanistic delineation to biomarker discovery. Genomics Proteomics Bioinformatics 19, 191–207 (2021).
Patti, G. J., Yanes, O. & Siuzdak, G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
Kishton, R. J., Sukumar, M. & Restifo, N. P. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab. 26, 94–109 (2017).
Fultang, L. et al. Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood 136, 1155–1160 (2020).
Hao, M. et al. Combination of metabolic intervention and T cell therapy enhances solid tumor immunotherapy. Sci. Transl. Med. 12, eaaz6667 (2020).
Hirabayashi, K. et al. Dual-targeting CAR-T cells with optimal co-stimulation and metabolic fitness enhance antitumor activity and prevent escape in solid tumors. Nat. Cancer 2, 904–918 (2021).
Vardhana, S. A. et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21, 1022–1033 (2020).
Gemta, L. F. et al. Impaired enolase 1 glycolytic activity restrains effector functions of tumor-infiltrating CD8+ T cells. Sci. Immunol. 4, eaap9520 (2019).
Shen, L. et al. Metabolic reprogramming by ex vivo glutamine inhibition endows CAR-T cells with less-differentiated phenotype and persistent antitumor activity. Cancer Lett. 538, 215710 (2022).
Xu, J. et al. Metabolic flux analysis and fluxomics-driven determination of reaction free energy using multiple isotopes. Curr. Opin. Biotechnol. 64, 151–160 (2020).
Liang, L., Sun, F., Wang, H. & Hu, Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol. Ther. 224, 107827 (2021).
Kawalekar, O. U. et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016).
Van Bruggen, J. A. C. et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood 134, 44–58 (2019).
Luo, M. et al. Race is a key determinant of the human intratumor microbiome. Cancer Cell https://doi.org/10.1016/j.ccell.2022.08.007 (2022).
Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, eabc4552 (2021).
Vétizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Spencer, C. N. et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 374, 1632–1640 (2021).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Smith, M. et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 28, 713–723 (2022).
Matson, V., Chervin, C. S. & Gajewski, T. F. Cancer and the microbiome—influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology 160, 600–613 (2021).
Moffitt, J. R., Lundberg, E. & Heyn, H. The emerging landscape of spatial profiling technologies. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00515-3 (2022).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 22, 627–644 (2021).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Kuett, L. et al. Three-dimensional imaging mass cytometry for highly multiplexed molecular and cellular mapping of tissues and the tumor microenvironment. Nat. Cancer 3, 122–133 (2022).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Schulz, D. et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 6, 25–36.e5 (2018).
Hoch, T. et al. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 7, eabk1692 (2022).
Hartmann, F. J. et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 39, 186–197 (2021).
Lee, W. D., Mukha, D., Aizenshtein, E. & Shlomi, T. Spatial-fluxomics provides a subcellular-compartmentalized view of reductive glutamine metabolism in cancer cells. Nat. Commun. 10, 1351 (2019).
Avila Cobos, F., Alquicira-Hernandez, J., Powell, J. E., Mestdagh, P. & De Preter, K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 11, 5650 (2020).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
Black, J. R. M. & McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer 21, 379–392 (2021).
Orlando, E. J. et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med. 24, 1504–1506 (2018).
Sammut, S.-J. et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601, 623–629 (2022).
Kang, M., Ko, E. & Mersha, T. B. A roadmap for multi-omics data integration using deep learning. Brief. Bioinform. 23, bbab454 (2022).
Goecks, J., Jalili, V., Heiser, L. M. & Gray, J. W. How machine learning will transform biomedicine. Cell 181, 92–101 (2020).
Miao, Z., Humphreys, B. D., McMahon, A. P. & Kim, J. Multi-omics integration in the age of million single-cell data. Nat. Rev. Nephrol. 17, 710–724 (2021).
Wei, J., Han, X., Bo, J. & Han, W. Target selection for CAR-T therapy. J. Hematol. Oncol. 12, 62 (2019).
Larson, R. C. & Maus, M. V. Recent advances and discoveries in the mechanisms and functions of CAR T cells. Nat. Rev. Cancer 21, 145–161 (2021).
Bosse, K. R. et al. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell 32, 295–309.e12 (2017).
Yarmarkovich, M. et al. Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs. Nature 599, 477–484 (2021).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Hsiue, E. H. C. et al. Targeting a neoantigen derived from a common TP53 mutation. Science 371, eabc8697 (2021).
Thorsson, V. et al. The immune landscape of cancer. Immunity 48, 812–830.e14 (2018).
Wu, J. et al. TSNAdb: a database for tumor-specific neoantigens from immunogenomics data analysis. Genomics Proteomics Bioinformatics 16, 276–282 (2018).
Dannenfelser, R. et al. Discriminatory power of combinatorial antigen recognition in cancer T cell therapies. Cell Syst. 11, 215–228.e5 (2020).
Kloss, C. C., Condomines, M., Cartellieri, M., Bachmann, M. & Sadelain, M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 31, 71–75 (2013).
Roybal, K. T. et al. Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell 167, 419–432.e16 (2016).
Fedorov, V. D., Themeli, M. & Sadelain, M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med. 5, 215ra172 (2013).
Cho, J. H., Collins, J. J. & Wong, W. W. Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell 173, 1426–1438.e11 (2018).
Cho, J. H. et al. Engineering advanced logic and distributed computing in human CAR immune cells. Nat. Commun. 12, 792 (2021).
Atilla, P. A. et al. Combinatorial antigen targeting strategies for acute leukemia: application in myeloid malignancy. Cytotherapy 24, 282–290 (2022).
Liu, F. et al. First-in-human CLL1-CD33 compound CAR T cell therapy induces complete remission in patients with refractory acute myeloid leukemia: update on phase 1 clinical trial. Blood 132, 901–901 (2018).
Spiegel, J. Y. et al. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: a phase 1 trial. Nat. Med. 27, 1419–1431 (2021).
Fernández de Larrea, C. et al. Defining an optimal dual-targeted CAR T-cell therapy approach simultaneously targeting BCMA and GPRC5D to prevent BCMA escape–driven relapse in multiple myeloma. Blood Cancer Discov. 1, 146–154 (2020).
Han, X. et al. Construction of a human cell landscape at single-cell level constructing an HCL using microwell-seq. Nature 581, 303–309 (2020).
He, S. et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 21, 294 (2020).
Tabula Sapiens Consortium et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Eraslan, G. et al. Single-nucleus cross-tissue molecular reference maps to decipher disease gene function. Science 376, eabl4290 (2022).
Suo, C. et al. Mapping the developing human immune system across organs. Science 376, eabo0510 (2022).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Bausch-Fluck, D. et al. The in silico human surfaceome. Proc. Natl Acad. Sci. USA 115, E10988–E10997 (2018).
Chen, N., Li, X., Chintala, N. K., Tano, Z. E. & Adusumilli, P. S. Driving CARs on the uneven road of antigen heterogeneity in solid tumors. Curr. Opin. Immunol. 51, 103–110 (2018).
Arcangeli, S. et al. CAR T cell manufacturing from naive/stem memory T lymphocytes enhances antitumor responses while curtailing cytokine release syndrome. J. Clin. Invest. 132, e150807 (2022).
Turtle, C. J. et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 126, 2123–2138 (2016).
Bai, Z. et al. Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci. Adv. 8, eabj2820 (2022).
Larson, R. C. et al. CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature 604, 563–570 (2022).
Seo, H. et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22, 983–995 (2021).
Tang, J. P. et al. Hypophosphatemia due to increased effector cell metabolic activity is associated with neurotoxicity symptoms in CD19-targeted CAR T-cell therapy. Cancer Immunol. Res. 10, 1433–1440 (2022).
Nobles, C. L. et al. CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J. Clin. Invest. 130, 673–685 (2020).
Cappell, K. M. & Kochenderfer, J. N. A comparison of chimeric antigen receptors containing CD28 versus 4-1BB costimulatory domains. Nat. Rev. Clin. Oncol. 18, 715–727 (2021).
Lemoine, J., Ruella, M. & Houot, R. Born to survive: how cancer cells resist CAR T cell therapy. J. Hematol. Oncol. 14, 199 (2021).
Lemoine, J., Ruella, M. & Houot, R. Overcoming intrinsic resistance of cancer cells to CAR T-cell killing. Clin. Cancer Res. 27, 6298–6306 (2021).
Plaks, V. et al. CD19 target evasion as a mechanism of relapse in large B-cell lymphoma treated with axicabtagene ciloleucel. Blood 138, 1081–1085 (2021).
Rabilloud, T. et al. Single-cell profiling identifies pre-existing CD19-negative subclones in a B-ALL patient with CD19-negative relapse after CAR-T therapy. Nat. Commun. 12, 865 (2021).
Asnani, M. et al. Retention of CD19 intron 2 contributes to CART-19 resistance in leukemias with subclonal frameshift mutations in CD19. Leukemia 34, 1202–1207 (2020).
Jacoby, E. et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat. Commun. 7, 12320 (2016).
Gardner, R. et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016).
Ruella, M. et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 24, 1499–1503 (2018).
Upadhyay, R. et al. A critical role for FAS-mediated off-target tumor killing in T-cell immunotherapy. Cancer Discov. 11, 599–613 (2021).
Monti, S. et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell 22, 359–372 (2012).
Jain, M. D. et al. Tumor interferon signaling and suppressive myeloid cells are associated with CAR T-cell failure in large B-cell lymphoma. Blood 137, 2621–2633 (2021).
Kankeu Fonkoua, L. A., Sirpilla, O., Sakemura, R., Siegler, E. L. & Kenderian, S. S. CAR T cell therapy and the tumor microenvironment: current challenges and opportunities. Mol. Ther. Oncolytics 25, 69–77 (2022).
Binnewies, M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018).
Britten, C. M., Shalabi, A. & Hoos, A. Industrializing engineered autologous T cells as medicines for solid tumours. Nat. Rev. Drug Discov. 20, 476–488 (2021).
Xu, N. et al. STING agonist promotes CAR T cell trafficking and persistence in breast cancer. J. Exp. Med. 218, e20200844 (2021).
Zhang, X. et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 12, 5291 (2021).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Adusumilli, P. S. et al. A phase I trial of regional mesothelin-targeted CAR T-cell therapy in patients with malignant pleural disease, in combination with the anti-PD-1 agent pembrolizumab. Cancer Discov. 11, 2748–2763 (2021).
Rafiq, S. et al. Targeted delivery of a PD-1-blocking scFV by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 36, 847–858 (2018).
Cherkassky, L. et al. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Invest. 126, 3130–3144 (2016).
Choi, B. D. et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunother. Cancer 7, 304 (2019).
Yamamoto, T. N. et al. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. J. Clin. Invest. 129, 1551–1565 (2019).
Avanzi, M. P. et al. Engineered tumor-targeted T cells mediate enhanced anti-tumor efficacy both directly and through activation of the endogenous immune system. Cell Rep. 23, 2130–2141 (2018).
Luu, M. et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 12, 4077 (2021).
Ellis, G. I., Sheppard, N. C. & Riley, J. L. Genetic engineering of T cells for immunotherapy. Nat. Rev. Genet. 22, 427–447 (2021).
Brudno, J. N. & Kochenderfer, J. N. Recent advances in CAR T-cell toxicity: mechanisms, manifestations and management. Blood Rev. 34, 45–55 (2019).
Kennedy, L. B. & Salama, A. K. S. A review of cancer immunotherapy toxicity. CA Cancer J. Clin. 70, 86–104 (2020).
Dougan, M., Luoma, A. M., Dougan, S. K. & Wucherpfennig, K. W. Understanding and treating the inflammatory adverse events of cancer immunotherapy. Cell 184, 1575–1588 (2021).
Morris, E. C., Neelapu, S. S., Giavridis, T. & Sadelain, M. Cytokine release syndrome and associated neurotoxicity in cancer immunotherapy. Nat. Rev. Immunol. 22, 85–96 (2022).
Teachey, D. T. et al. Identification of predictive biomarkers for cytokine release syndrome after chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Cancer Discov. 6, 664–679 (2016).
Gill, S. & Brudno, J. N. CAR T-cell therapy in hematologic malignancies: clinical role, toxicity, and unanswered questions. Am. Soc. Clin. Oncol. Educ. B 41, e246–e265 (2021).
Depil, S., Duchateau, P., Grupp, S. A., Mufti, G. & Poirot, L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 19, 185–199 (2020).
Bai, Z. et al. Single-cell multiomics dissection of basal and antigen-specific activation states of CD19-targeted CAR T cells. J. Immunother. Cancer 9, e002328 (2021).
Davis-Marcisak, E. F. et al. From bench to bedside: single-cell analysis for cancer immunotherapy. Cancer Cell 39, 1062–1080 (2021).
Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Andrews, T. S., Kiselev, V. Y., McCarthy, D. & Hemberg, M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat. Protoc. 16, 2749–2764 (2021).
Guo, S., Zhang, C. & Le, A. The limitless applications of single-cell metabolomics. Curr. Opin. Biotechnol. 71, 115–122 (2021).
Lloréns-Rico, V., Simcock, J. A., Huys, G. R. B. & Raes, J. Single-cell approaches in human microbiome research. Cell 185, 2725–2738 (2022).
Johnson, D. B. et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin. Cancer Res. 24, 5250–5260 (2018).
Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020).
Jing, Y., Yang, J., Johnson, D. B., Moslehi, J. J. & Han, L. Harnessing big data to characterize immune-related adverse events. Nat. Rev. Clin. Oncol. 19, 269–280 (2022).
Reel, P. S., Reel, S., Pearson, E., Trucco, E. & Jefferson, E. Using machine learning approaches for multi-omics data analysis: a review. Biotechnol. Adv. 49, 107739 (2021).
Levine, L. S. et al. Single-cell analysis by mass cytometry reveals metabolic states of early-activated CD8+ T cells during the primary immune response. Immunity 54, 829–844.e5 (2021).
Acknowledgements
The work of the authors is supported by the US NIH (grants R01HG011633 and R01CA262623 to L.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. M.R.G. is a Scholar of the Leukaemia and Lymphoma Society. The authors regret that page limitations have prevented them from including all the relevant studies in this Review. Draft figures for this manuscript were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to all aspects of the preparation of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.R.G. has stock ownership interest in KDAc Therapeutics, receives funding from Abbvie, Allogene, Kite/Gilead and Sanofi, and has received honoraria from Tessa Therapeutics. J.Y., Y.C., Y.J. and L.H. declare no competing interests.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks M. Maus, who co-reviewed with N. Haradhvala; R. Fan, who co-reviewed with Z. Bai; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Antigen Explorer: https://antigen.princeton.edu/
CARTSC: https://hanlab.tamhsc.edu/CARTSC/
ClinicalTrials.gov: https://clinicaltrials.gov/ct2/home
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Chen, Y., Jing, Y. et al. Advancing CAR T cell therapy through the use of multidimensional omics data. Nat Rev Clin Oncol 20, 211–228 (2023). https://doi.org/10.1038/s41571-023-00729-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-023-00729-2
This article is cited by
-
Targeting immunogenic cell stress and death for cancer therapy
Nature Reviews Drug Discovery (2024)
-
Recent Advances and Challenges in Cancer Treatment with Car T Cell Therapy: A Novel Anti-cancer Strategy
BioNanoScience (2024)
-
Novel research and future prospects of artificial intelligence in cancer diagnosis and treatment
Journal of Hematology & Oncology (2023)
-
Radiation therapy improves CAR T cell activity in acute lymphoblastic leukemia
Cell Death & Disease (2023)
-
Immunotherapy in hematologic malignancies: achievements, challenges and future prospects
Signal Transduction and Targeted Therapy (2023)