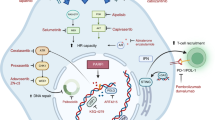
Abstract
Developing novel targeted anticancer therapies is a major goal of current research. The use of poly(ADP-ribose) polymerase (PARP) inhibitors in patients with homologous recombination-deficient tumours provides one of the best examples of a targeted therapy that has been successfully translated into the clinic. The success of this approach has so far led to the approval of four different PARP inhibitors for the treatment of several types of cancers and a total of seven different compounds are currently under clinical investigation for various indications. Clinical trials have demonstrated promising response rates among patients receiving PARP inhibitors, although the majority will inevitably develop resistance. Preclinical and clinical data have revealed multiple mechanisms of resistance and current efforts are focused on developing strategies to address this challenge. In this Review, we summarize the diverse processes underlying resistance to PARP inhibitors and discuss the potential strategies that might overcome these mechanisms such as combinations with chemotherapies, targeting the acquired vulnerabilities associated with resistance to PARP inhibitors or suppressing genomic instability.
Key points
-
BRCA1/2-deficient cells are extremely vulnerable to PARP inhibition; this finding has led to the successful clinical development of PARP inhibitors for the treatment of patients with BRCA1/2-mutant cancers.
-
The use of PARP inhibitors is currently being expanded beyond patients with ovarian and breast cancers to those with homologous recombination-deficient tumours of other histologies such as prostate or pancreatic cancers.
-
Resistance to PARP inhibitors occurs through three main mechanisms: drug target-related resistance, restoration of homologous recombination and restoration of replication fork stability.
-
Current approaches to overcome resistance are focused on combining PARP inhibition with other inhibitors of the DNA damage response, immune-checkpoint inhibition or targeted therapies.
-
Targeting the acquired vulnerabilities of PARP inhibitor-resistant tumours might selectively kill unresponsive cancer cells.
-
Preventing the development of genomic instability in DNA repair-deficient cancers by inhibiting the activity of the error-prone polymerase POLθ might be a promising strategy to target a known vulnerability of these tumours.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Bai, P. & Cantó, C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 16, 290–295 (2012).
Hanzlikova, H. et al. The importance of poly(ADP-ribose) polymerase as a sensor of unligated okazaki fragments during DNA replication. Mol. Cell 71, 319–331.e3 (2018).
Ray Chaudhuri, A. & Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017).
Chen, C., Feng, W., Lim, P. X., Kass, E. M. & Jasin, M. Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2, 313–336 (2017).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Schlacher, K. et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Patch, A. M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015).
Armenia, J. et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018).
Pritchard, C. C. et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 375, 443–453 (2016).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Ashworth, A. & Lord, C. J. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 15, 564–576 (2018).
Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R. & Bishop, D. K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 (2000).
Treszezamsky, A. D. et al. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 67, 7078–7081 (2007).
Evers, B. et al. A high-throughput pharmaceutical screen identifies compounds with specific toxicity against BRCA2-deficient tumors. Clin. Cancer Res. 16, 99–108 (2010).
Slade, D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 34, 360–394 (2020).
Pilié, P. G., Tang, C., Mills, G. B. & Yap, T. A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 16, 81–104 (2019).
Mateo, J. et al. A decade of clinical development of PARP inhibitors in perspective. Ann. Oncol. 30, 1437–1447 (2019).
Curtin, N. J. & Szabo, C. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat. Rev. Drug Discov. 19, 711–736 (2020).
Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 5, 387–393 (2011).
Ström, C. E. et al. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 39, 3166–3175 (2011).
Patel, A. G., Sarkaria, J. N. & Kaufmann, S. H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl Acad. Sci. USA 108, 3406–3411 (2011).
Zahradka, P. & Ebisuzaki, K. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127, 579–585 (1982).
Murai, J. et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014).
Murai, J. & Pommier, Y. PARP trapping beyond homologous recombination and platinum sensitivity in cancers. Annu. Rev. Cancer Biol. 3, 131–150 (2019).
Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012).
Shen, Y. et al. BMN673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 19, 5003–5015 (2013).
Zandarashvili, L. et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367 (2020).
Brown, J. S., Kaye, S. B. & Yap, T. A. PARP inhibitors: the race is on. Br. J. Cancer 114, 713–715 (2016).
Carney, B. et al. Target engagement imaging of PARP inhibitors in small-cell lung cancer. Nat. Commun. 9, 176 (2018).
Pommier, Y., O’Connor, M. J. & De Bono, J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps17 (2016).
LaFargue, C. J., Dal Molin, G. Z., Sood, A. K. & Coleman, R. L. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol. 20, e15–e28 (2019).
Zimmermann, M. et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285–289 (2018).
Schoonen, P. M. et al. Progression through mitosis promotes PARP inhibitor-induced cytotoxicity in homologous recombination-deficient cancer cells. Nat. Commun. 8, 15981 (2017).
Maya-Mendoza, A. et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284 (2018).
Kaufman, B. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250 (2015).
Kim, G. et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 21, 4257–4261 (2015).
Ledermann, J. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian. Cancer N. Engl. J. Med. 366, 1382–1392 (2012).
Balasubramaniam, S. et al. FDA approval summary: rucaparib for the treatment of patients with deleterious BRCA mutation–associated advanced ovarian cancer. Clin. Cancer Res. 23, 7165–7170 (2017).
Swisher, E. M. et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 18, 75–87 (2017).
Kristeleit, R. S. et al. Clinical activity of the poly(ADP-ribose) polymerase (PARP) inhibitor rucaparib in patients (pts) with high-grade ovarian carcinoma (HGOC) and a BRCA mutation (BRCAmut): analysis of pooled data from Study 10 (parts 1, 2a, and 3) and ARIEL2 (parts 1 and 2). Ann. Oncol. 27, 296–312 (2016).
Moore, K. N. et al. QUADRA: a phase 2, open-label, single-arm study to evaluate niraparib in patients (pts) with relapsed ovarian cancer (ROC) who have received≥3 prior chemotherapy regimens. J. Clin. Oncol. 36, 5514–5514 (2018).
Moore, K. N. et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 20, 636–648 (2019).
Ledermann, J. et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 15, 852–861 (2014).
Pujade-Lauraine, E. et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1274–1284 (2017).
Mirza, M. R. et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 375, 2154–2164 (2016).
Coleman, R. L. et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1949–1961 (2017).
Moore, K. et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 379, 2495–2505 (2018).
González-Martín, A. et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 381, 2391–2402 (2019).
Robson, M. et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533 (2017).
Litton, J. K. et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 379, 753–763 (2018).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
de Bono, J. et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 382, 2091–2102 (2020).
Ganesan, S. & Garber, J. Poly (ADP-Ribose) polymerase inhibitor activity in prostate cancers harboring mutations in DNA repair genes: who benefits? JCO Precis. Oncol. https://doi.org/10.1200/PO.20.00269 (2020).
Abida, W. et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J. Clin. Oncol. 38, 3763–3772 (2020).
Lickliter, J. D. et al. A phase I dose-escalation study of BGB-290, a novel PARP1/2 selective inhibitor in patients with advanced solid tumors. J. Clin. Oncol. 34, e17049 (2016).
Lickliter, J. et al. Dose escalation/expansion study to investigate the safety, pharmacokinetics, food effect, and antitumor activity of BGB-290 in patients with advanced solid tumors. Ann. Oncol. 28, v123 (2017).
Wang, L. et al. Pharmacologic characterization of fluzoparib, a novel poly(ADP-ribose) polymerase inhibitor undergoing clinical trials. Cancer Sci. 110, 1064–1075 (2019).
Ledermann, J. A. et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 17, 1579–1589 (2016).
Friedlander, M. L. et al. 234O Maintenance olaparib for patients (pts) with newly diagnosed, advanced ovarian cancer (OC) and a BRCA mutation (BRCAm): 5-year (y) follow-up (f/u) from SOLO1. Ann. Oncol. 31 (Suppl. 3), S1334 (2020).
Andrés, P. et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 22, 620–631 (2021).
Coleman, R. L. et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N. Engl. J. Med. 381, 2403–2415 (2019).
Ray-Coquard, I. et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N. Engl. J. Med. 381, 2416–2428 (2019).
McCabe, N. et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115 (2006).
Hodgson, D. R. et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 119, 1401–1409 (2018).
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015).
Abida, W. et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase II TRITON2 Study. Clin. Cancer Res. 26, 2487–2496 (2020).
Tung, N. M. et al. TBCRC 048: a phase II study of olaparib monotherapy in metastatic breast cancer patients with germline or somatic mutations in DNA damage response (DDR) pathway genes (Olaparib Expanded). J. Clin. Oncol. 38, 1002 (2020).
Grellety, T. et al. Dramatic response to PARP inhibition in a PALB2-mutated breast cancer: moving beyond BRCA. Ann. Oncol. 31, 822–823 (2020).
Rahman, N. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39, 165–167 (2007).
Kondrashova, O. et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7, 984–998 (2017).
Bang, Y. J. et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1637–1651 (2017).
Shen, J. et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5, 752–767 (2015).
Ismail, I. H. et al. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 74, 4282–4294 (2014).
Parrotta, R. et al. A novel BRCA1-associated protein-1 isoform affects response of mesothelioma cells to drugs impairing BRCA1-mediated DNA repair. J. Thorac. Oncol. 12, 1309–1319 (2017).
Sulkowski, P. L. et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 50, 1086–1092 (2018).
Sulkowski, P. L. et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 582, 586–591 (2020).
Pietanza, M. C. et al. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018).
Sen, T., Gay, C. M. & Byers, L. A. Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl. Lung Cancer Res. 7, 50–68 (2018).
Miller, R. E. et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 31, 1606–1622 (2020).
Jaspers, J. E. et al. BRCA2-deficient sarcomatoid mammary tumors exhibit multidrug resistance. Cancer Res. 75, 732–741 (2015).
Rottenberg, S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17079–17084 (2008).
Vaidyanathan, A. et al. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 115, 431–441 (2016).
Pettitt, S. J. et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018).
Pettitt, S. J. et al. A genetic screen using the piggybac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PLoS One 8, 1–10 (2013).
Gogola, E. et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 33, 1078–1093.e12 (2018).
Edwards, S. L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Sakai, W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008).
Ter Brugge, P. et al. Mechanisms of therapy resistance in patient-derived xenograft models of brca1-deficient breast cancer. J. Natl Cancer Inst. 108, 1–12 (2016).
Kondrashova, O. et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 9, 3970 (2018).
Afghahi, A. et al. Tumor BRCA1 reversion mutation arising during neoadjuvant platinum-based chemotherapy in triple-negative breast cancer is associated with therapy resistance. Clin. Cancer Res. 23, 3365–3370 (2017).
Weigelt, B. et al. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin. Cancer Res. 23, 6708–6720 (2017).
Barber, L. J. et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 229, 422–429 (2013).
Norquist, B. et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 29, 3008–3015 (2011).
Domchek, S. M. Reversion mutations with clinical use of PARP inhibitors: many genes, many. versions. Cancer Discov. 7, 937–939 (2017).
Lin, K. K. et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 9, 210–219 (2019).
Pishvaian, M. J. et al. BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. Br. J. Cancer 116, 1021–1026 (2017).
Quigley, D. et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 7, 999–1005 (2017).
Goodall, J. et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7, 1006–1017 (2017).
Pettitt, S. J. et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 10, 1475–1488 (2020).
Ang, J. E. et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: A multi-institutional study. Clin. Cancer Res. 19, 5485–5493 (2013).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 (2010).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Cao, L. et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541 (2009).
Di Virgilio, M. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 (2013).
Feng, L., Fong, K. W., Wang, J., Wang, W. & Chen, J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288, 11135–11143 (2013).
Escribano-Díaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013).
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013).
Zimmermann, M., Lottersberger, F., Buonomo, S. B., Sfeir, A. & de Lange, T. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science 339, 700–704 (2013).
Xu, G. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015).
Boersma, V. et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521, 537–540 (2015).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018).
Dev, H. et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20, 954–965 (2018).
Ghezraoui, H. et al. 53BP1 cooperation with the REV7–shieldin complex underpins DNA structure-specific NHEJ. Nature 560, 122–127 (2018).
Gupta, R. et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988.e23 (2018).
Gao, S. et al. An OB-fold complex controls the repair pathways for DNA double-strand breaks. Nat. Commun. 9, 3925 (2018).
Findlay, S. et al. SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. 37, e100158 (2018).
Tomida, J. et al. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 37, e99543 (2018).
Jaspers, J. E. et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1 -mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013).
Cruz, C. et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 29, 1203–1210 (2018).
Waks, A. G. et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann. Oncol. 31, 590–598 (2020).
Johnson, N. et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl Acad. Sci. USA 110, 17041–17046 (2013).
Barazas, M. et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep. 23, 2107–2118 (2018).
Mirman, Z. et al. 53BP1–RIF1–shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560, 112–116 (2018).
Tkáč, J. et al. HELB is a feedback inhibitor of DNA end resection. Mol. Cell 61, 405–418 (2016).
He, Y. J. et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 563, 522–526 (2018).
Becker, J. R. et al. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat. Commun. 9, 5406 (2018).
Olivieri, M. et al. A genetic map of the response to DNA damage in human cells. Cell 182, 481–496.e21 (2020).
Francica, P. et al. Functional radiogenetic profiling implicates ERCC6L2 in Non-homologous end joining. Cell Rep. 32, 108068 (2020).
Drané, P. et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function HHS Public Access. Nature 543, 211–216 (2017).
Clairmont, C. S. et al. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 22, 87–96 (2020).
Choi, Y. E. et al. Platinum and PARP inhibitor resistance due to overexpression of MicroRNA-622 in BRCA1-mutant ovarian cancer. Cell Rep. 14, 429–439 (2016).
Ray Chaudhuri, A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016).
Ying, S., Hamdy, F. C. & Helleday, T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72, 2814–2821 (2012).
Guillemette, S. et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 29, 489–494 (2015).
Taglialatela, A. et al. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell 68, 414–430.e8 (2017).
Kolinjivadi, A. M. et al. Smarcal1-mediated fork reversal triggers mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell 67, 867–881.e7 (2017).
Dungrawala, H. et al. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell 67, 374–386.e5 (2017).
Rondinelli, B. et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 19, 1371–1378 (2017).
Lai, X. et al. MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2-deficient cells. Nat. Commun. 8, 15983 (2017).
Lemaçon, D. et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8, 860 (2017).
Ding, X. et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 7, 12425 (2016).
Zoppoli, G. et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl Acad. Sci. USA 109, 15030–15035 (2012).
Stewart, C. A. et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8, 28575–28587 (2017).
Lok, B. H. et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 23, 523–535 (2017).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Murai, J. et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 7, 76534–76550 (2016).
Murai, J. et al. SLFN11 blocks stressed replication forks independently of ATR. Mol. Cell 69, 371–384.e6 (2018).
Zhou, Y., Caron, P., Legube, G. & Paull, T. T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42, e19 (2014).
Polato, F. et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 211, 1027–1036 (2014).
Reczek, C. R., Szabolcs, M., Stark, J. M., Ludwig, T. & Baer, R. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 201, 693–707 (2013).
Cruz-García, A., López-Saavedra, A. & Huertas, P. BRCA1 accelerates CtIP-ediated DNA-end resection. Cell Rep. 9, 451–459 (2014).
Nacson, J. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep. 24, 3513–3527.e7 (2018).
Nakada, S., Yonamine, R. M. & Matsuo, K. RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res. 72, 4974–4983 (2012).
Luijsterburg, M. S. & et al. A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation.eLife 6, e20922 (2017).
Zong, D. et al. BRCA1 haploinsufficiency is masked by RNF168-mediated chromatin ubiquitylation. Mol. Cell 73, 1267–1281.e7 (2019).
Belotserkovskaya, R. et al. PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1. Nat. Commun. 11, 819 (2020).
Callen, E. et al. 53BP1 enforces distinct pre- and post-resection blocks on homologous recombination. Mol. Cell 77, 26–38.e7 (2020).
Rijkers, T. et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429 (1998).
Yamaguchi-Iwai, Y. et al. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol. 18, 6430–6435 (1998).
Feng, Z. et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl Acad. Sci. USA 108, 686–691 (2011).
Lok, B. H., Carley, A. C., Tchang, B. & Powell, S. N. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32, 3552–3558 (2013).
Malacaria, E. et al. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat. Commun. 10, 1412 (2019).
Sotiriou, S. K. et al. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell 64, 1127–1134 (2016).
Hengel, S. R., Spies, M. A. & Spies, M. Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem. Biol. 24, 1101–1119 (2017).
Sullivan-Reed, K. et al. Simultaneous targeting of PARP1 and RAD52 triggers dual synthetic lethality in BRCA-deficient tumor cells. Cell Rep. 23, 3127–3136 (2018).
Mirza, M. R. et al. Niraparib plus bevacizumab versus niraparib alone for platinum-sensitive recurrent ovarian cancer (NSGO-AVANOVA2/ENGOT-ov24): a randomised, phase 2, superiority trial. Lancet Oncol. 20, 1409–1419 (2019).
Yap, T. A. et al. Phase I trial of the parp inhibitor olaparib and Akt inhibitor capivasertib in patients with brca1/2-and non–brca1/2-mutant cancers. Cancer Discov. 10, 1528–1543 (2020).
Kurnit, K. C. et al. Abstract CT020: phase I dose escalation of olaparib (PARP inhibitor) and selumetinib (MEK inhibitor) combination in solid tumors with Ras pathway alterations. Cancer Res. 79 (Suppl. 13), CT020 (2019).
Sun, C. et al. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell 33, 401–416.e8 (2018).
Yang, L. et al. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl. Med. 9, eaal1645 (2017).
Karakashev, S. et al. BET bromodomain inhibition synergizes with PARP inhibitor in epithelial ovarian cancer. Cell Rep. 21, 3398–3405 (2017).
Min, A. et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 17, 33 (2015).
Konstantinopoulos, P. A., Wilson, A. J., Saskowski, J., Wass, E. & Khabele, D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol. Oncol. 133, 599–606 (2014).
Chao, O. S. & Goodman, O. B. Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol. Cancer Res. 12, 1755–1766 (2014).
Marijon, H. et al. Co-targeting poly(ADP-ribose) polymerase (PARP) and histone deacetylase (HDAC) in triple-negative breast cancer: Higher synergism in BRCA mutated cells. Biomed. Pharmacother. 99, 543–551 (2018).
Johnson, N. et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat. Med. 17, 875–882 (2011).
Johnson, S. F. et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 17, 2367–2381 (2016).
Joshi, P. M., Sutor, S. L., Huntoon, C. J. & Karnitz, L. M. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 289, 9247–9253 (2014).
Bajrami, I. et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287–297 (2014).
Choi, Y. E. et al. Sublethal concentrations of 17-AAG suppress homologous recombination DNA repair and enhance sensitivity to carboplatin and olaparib in HR proficient ovarian cancer cells. Oncotarget 5, 2678–2687 (2014).
Jiang, J. et al. Ganetespib overcomes resistance to PARP inhibitors in breast cancer by targeting core proteins in the DNA repair machinery. Invest. N. Drugs 35, 251–259 (2017).
Krawczyk, P. M. et al. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc. Natl Acad. Sci. USA 108, 9851–9856 (2011).
van den Tempel, N. et al. Heat-induced BRCA2 degradation in human tumours provides rationale for hyperthermia-PARP-inhibitor combination therapies. Int. J. Hyperth. 34, 407–414 (2018).
Lu, Y., Chu, A., Turker, M. S. & Glazer, P. M. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol. Cell. Biol. 31, 3339–3350 (2011).
Yazinski, S. A. et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017).
Kim, H. et al. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin. Cancer Res. 23, 3097–3108 (2017).
Kim, H. et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat. Commun. 11, 3726 (2020).
Parsels, L. A. et al. PARP1 trapping and DNA replication stress enhance radiosensitization with combined WEE1 and PARP inhibitors. Mol. Cancer Res. 16, 222–232 (2018).
Lallo, A. et al. The combination of the PARP inhibitor olaparib and the WEE1 inhibitor AZD1775 as a new therapeutic option for small cell lung cancer. Clin. Cancer Res. 24, 5153–5164 (2018).
Fang, Y. et al. Sequential therapy with PARP and WEE1 inhibitors minimizes toxicity while maintaining efficacy. Cancer Cell 35, 851–867.e7 (2019).
Pilié, P. G., Gay, C. M., Byers, L. A., O’Connor, M. J. & Yap, T. A. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin. Cancer Res. 25, 3759–3771 (2019).
Houtkooper, R. H., Cantó, C., Wanders, R. J. & Auwerx, J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194–223 (2010).
Virag, L. The therapeutic potential of poly(ADP-Ribose) polymerase inhibitors. Pharmacol. Rev. 54, 375–429 (2002).
Eliasson, M. J. L. et al. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med. 3, 1089–1095 (1997).
Zong, W.-X. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes. Dev. 18, 1272–1282 (2004).
Bajrami, I. et al. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol. Med. 4, 1087–1096 (2012).
Tateishi, K. et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 28, 773–784 (2015).
Lu, Y. et al. Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mediated DNA repair. Cancer Res. 77, 1709–1718 (2017).
Sulkowski, P. L. et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 9, eaal2463 (2017).
Bian, C. et al. NADP+is an endogenous PARP inhibitor in DNA damage response and tumor suppression. Nat. Commun. 10, 693 (2019).
Strickland, K. C. et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 7, 13587–13598 (2016).
Parkes, E. E. et al. Activation of STING-dependent innate immune signaling by s-phase-specific DNA damage in breast cancer. J. Natl Cancer Inst. 109, 1–10 (2017).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Zhu, Q. et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 (2011).
Shen, J. et al. PARPI triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 79, 311–319 (2019).
Ding, L. et al. PARP inhibition elicits STING-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell Rep. 25, 2972–2980.e5 (2018).
Jiao, S. et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 23, 3711–3720 (2017).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Li, A. et al. Prospects for combining immune checkpoint blockade with PARP inhibition. J. Hematol. Oncol. 12, 98 (2019).
Friedlander, M. et al. Pamiparib in combination with tislelizumab in patients with advanced solid tumours: results from the dose-escalation stage of a multicentre, open-label, phase 1a/b trial. Lancet Oncol. 20, 1306–1315 (2019).
Domchek, S. M. et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol. 21, 1155–1164 (2020).
Konstantinopoulos, P. A. et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. 5, 1141–1149 (2019).
Leite de Oliveira, R., Wang, L. & Bernards, R. With great power comes great vulnerability. Mol. Cell. Oncol. 5, e1509488 (2018).
Ame, J.-C. et al. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 122, 1990–2002 (2009).
Callen, E. et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280 (2013).
Ward, I. M., Minn, K., van Deursen, J. & Chen, J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23, 2556–2563 (2003).
Barazas, M. et al. Radiosensitivity is an acquired vulnerability of PARPi-resistant BRCA1-deficient tumors. Cancer Res. 79, 452–460 (2019).
Eke, I. et al. 53BP1/RIF1 signaling promotes cell survival after multifractionated radiotherapy. Nucleic Acids Res. 48, 1314–1326 (2020).
Feng, F. Y. et al. Targeted radiosensitization with PARP1 inhibition: optimization of therapy and identification of biomarkers of response in breast cancer. Breast Cancer Res. Treat. 147, 81–94 (2014).
Murai, J. et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J. Pharmacol. Exp. Ther. 349, 408–416 (2014).
Tentori, L. et al. Poly(ADP-ribose) glycohydrolase inhibitor as chemosensitiser of malignant melanoma for temozolomide. Eur. J. Cancer 41, 2948–2957 (2005).
Nagashima, H. et al. Poly(ADP-ribose) glycohydrolase inhibition sequesters NAD+ to potentiate the metabolic lethality of alkylating chemotherapy in IDH-mutant tumor cells. Cancer Discov. 10, 1672–1689 (2020).
Michelena, J. et al. Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance. Nat. Commun. 9, 2678 (2018).
Fleury, H. et al. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence. Nat. Commun. 10, 2556 (2019).
Shen, S., Vagner, S. & Robert, C. Persistent cancer cells: the deadly survivors. Cell 183, 860–874 (2020).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015).
Mateos-Gomez, P. A. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Zhou, J., Gelot, C., Pantelidou, C. et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat Cancer 2, 598–610 (2021).
Wheeler, D. A. et al. Molecular features of cancers exhibiting exceptional responses to treatment. Cancer Cell 39, 38–53.e7 (2021).
Xiong, Y. et al. Pamiparib is a potent and selective PARP inhibitor with unique potential for the treatment of brain tumor. Neoplasia 22, 431–440 (2020).
European Medicines Agency. Lynparza Annex I. Eur. Med. Agency https://www.ema.europa.eu/en/medicines/human/EPAR/lynparza (2021).
European Medicines Agency. Rubraca Annex I. Eur. Med. Agency https://www.ema.europa.eu/en/medicines/human/EPAR/rubraca (2021).
European Medicines Agency. Talzenna Annex I. Eur. Med. Agency https://www.ema.europa.eu/en/medicines/human/EPAR/talzenna (2021).
European Medicines Agency. Zejula Annex 1. Eur Med. Agency https://www.ema.europa.eu/en/medicines/human/EPAR/zejula (2021).
Wahlberg, E. et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 30, 283–288 (2012).
Halford, S. E. R. et al. Results of the OPARATIC trial: a phase I dose escalation study of olaparib in combination with temozolomide (TMZ) in patients with relapsed glioblastoma (GBM). J. Clin. Oncol. 35, 2022 (2017).
Daniel, R. A. et al. Central nervous system penetration and enhancement of temozolomide activity in childhood medulloblastoma models by poly(ADP-ribose) polymerase inhibitor AG-014699. Br. J. Cancer 103, 1588–1596 (2010).
Sun, K. et al. A comparative pharmacokinetic study of PARP inhibitors demonstrates favorable properties for niraparib efficacy in preclinical tumor models. Oncotarget 9, 37080–37096 (2018).
Kizilbash, S. H. et al. Restricted delivery of talazoparib across the blood–brain barrier limits the sensitizing effects of PARP inhibition on temozolomide therapy in glioblastoma. Mol. Cancer Ther. 16, 2735–2746 (2017).
Pothuri, B. et al. Phase I and pharmacokinetic study of veliparib, a PARP inhibitor, and pegylated liposomal doxorubicin (PLD) in recurrent gynecologic cancer and triple negative breast cancer with long-term follow-up. Cancer Chemother. Pharmacol. 85, 741–751 (2020).
Werner, T. L. et al. Safety and pharmacokinetics of veliparib extended-release in patients with advanced solid tumors: a phase I study. Cancer Med. 7, 2360–2369 (2018).
Gibson, B. A. & Kraus, W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012).
Pascal, J. M. & Ellenberger, T. The rise and fall of poly(ADP-ribose): an enzymatic perspective. DNA Repair. 32, 10–16 (2015).
Amé, J.-C. et al. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 (1999).
Hanzlikova, H., Gittens, W., Krejcikova, K., Zeng, Z. & Caldecott, K. W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 45, 2546–2557 (2017).
Grundy, G. J. et al. PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B Glu2. Nat. Commun. 7, 12404 (2016).
Rulten, S. L. et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell 41, 33–45 (2011).
Langelier, M.-F., Riccio, A. A. & Pascal, J. M. NAR Breakthrough Article PARP-2 and PARP-3 are selectively activated by 5 phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 42, 7762–7775 (2014).
Loseva, O. et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 285, 8054–8060 (2010).
Mueller-Dieckmann, C. et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006).
Abplanalp, J. et al. Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 8, 2055 (2017).
Barkauskaite, E. et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun. 4, 2164 (2013).
Liu, C., Wu, J., Paudyal, S. C., You, Z. & Yu, X. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 41, 1698–1710 (2013).
Roy, R., Chun, J. & Powell, S. N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 (2012).
Harbeck, N. et al. Breast cancer. Nat. Rev. Dis. Prim. 5, 66 (2019).
Xia, B. et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 (2006).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Vidula, N. et al. Routine plasma-based genotyping to comprehensively detect germline, somatic, and reversion BRCA mutations among patients with advanced solid tumors. Clin. Cancer Res. 26, 2546–2555 (2020).
Acknowledgements
Work in J.J.’s laboratory was funded by the Oncode Institute, which is partly financed by the Dutch Cancer Society. The work of M.P.D. is supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 722729. The work of S.C.M. is supported by Boehringer Ingelheim Fonds. S.G. has received funding from the NCI (grants R01-CA243547, RO1-CA202752 and 5P30CA072720-21), US Department of Defence, the Breast Cancer Research Foundation, Hugs for Brady, the Val Skinner Foundation, the Gertrude Fogarty Trust and AHEPA.
Author information
Authors and Affiliations
Contributions
M.P.D. and S.C.M. made substantial contributions to researching data for this article and discussions of content. All authors contributed to writing the article and reviewing/editing of the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
J.J. has acted as a consultant of and received research support from Artios Pharma. S.G. has acted as a consultant of Foghorn Therapeutics, Foundation Medicine, Inspirata, Merck, Novartis, and Roche and has received research funding from M2Gen; his spouse is an employee of Merck and holds equity in Merck. The other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Clinical Oncology thanks G. Peng and I. Ray-Coquard for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dias, M.P., Moser, S.C., Ganesan, S. et al. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol 18, 773–791 (2021). https://doi.org/10.1038/s41571-021-00532-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-021-00532-x
This article is cited by
-
Drug resistance in ovarian cancer: from mechanism to clinical trial
Molecular Cancer (2024)
-
Subsequent management and outcomes after first-line PARP inhibitors progression in ovarian cancer patients
Journal of Ovarian Research (2024)
-
PARG-deficient tumor cells have an increased dependence on EXO1/FEN1-mediated DNA repair
The EMBO Journal (2024)
-
DNA double-strand break–capturing nuclear envelope tubules drive DNA repair
Nature Structural & Molecular Biology (2024)
-
Aryl hydrocarbon receptor suppresses STING-mediated type I IFN expression in triple-negative breast cancer
Scientific Reports (2024)