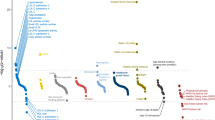
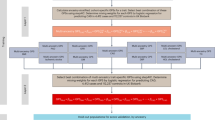
Abstract
Over the past decade, substantial progress has been made in the discovery of alleles contributing to the risk of coronary artery disease. In addition to providing causal insights into disease, these endeavours have yielded and enabled the refinement of polygenic risk scores. These scores can be used to predict incident coronary artery disease in multiple cohorts and indicate the clinical response to some preventive therapies in post hoc analyses of clinical trials. These observations and the widespread ability to calculate polygenic risk scores from direct-to-consumer and health-care-associated biobanks have raised many questions about responsible clinical adoption. In this Review, we describe technical and downstream considerations for the derivation and validation of polygenic risk scores and current evidence for their efficacy and safety. We discuss the implementation of these scores in clinical medicine for uses including risk prediction and screening algorithms for coronary artery disease, prioritization of patient subgroups that are likely to derive benefit from treatment, and efficient prospective clinical trial designs.
Key points
-
Genome-wide association studies demonstrate that multiple common genetic variants predispose individuals to coronary artery disease (CAD).
-
Polygenic risk scores (PRS) are singular, quantitative metrics for genetic susceptibility to a disease such as CAD.
-
The predictive performance of PRS for CAD is improved by incorporating evidence for association, linkage disequilibrium, anticipated functional impact and pleiotropy; trans-ancestry data improve the use of PRS in populations of diverse ancestry.
-
Post hoc analyses from completed clinical trials indicate that individuals with a high PRS for CAD derive the greatest relative and absolute benefit from LDL cholesterol-lowering strategies.
-
PRS for CAD could be used to identify individuals who would benefit from intensive lifestyle modification, imaging surveillance and early statin therapy.
-
PRS for CAD could be used to identify individuals at high risk for efficient clinical trial enrolment, and evidence of heterogeneous treatment benefit could be assessed through innovative trial designs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
White, P. D. Genes, the heart and destiny. N. Engl. J. Med. 256, 965–969 (1957).
Gertler, M. M., Garn, S. M. & White, P. D. Young candidates for coronary heart disease. JAMA 147, 621–625 (1951).
Marenberg, M. E., Risch, N., Berkman, L. F., Floderus, B. & de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 330, 1041–1046 (1994).
Natarajan, P. Polygenic risk scoring for coronary heart disease: the first risk factor. J. Am. Coll. Cardiol. 72, 1894–1897 (2018).
Inouye, M. et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J. Am. Coll. Cardiol. 72, 1883–1893 (2018).
Aragam, K. G. & Natarajan, P. Polygenic scores to assess atherosclerotic cardiovascular disease risk: clinical perspectives and basic implications. Cir. Res. 126, 1159–1177 (2020).
Nsengiman, J. et al. Enhanced linkage of a locus on chromosome 2 to premature coronary artery disease in the absence of hypercholesterolemia. Eur. J. Hum. Genet. 15, 313–319 (2007).
Khera, A. V. et al. Diagnostic yield of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J. Am. Coll. Cardiol. 67, 2578–2589 (2016).
Hu, P. et al. Prevalence of familial hypercholesterolemia among the general population and patients with atherosclerotic cardiovascular disease: a systematic review and meta-analysis. Circulation 141, 1742–1759 (2020).
Rader, D. J., Cohen, J. & Hobbs, H. H. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J. Clin. Invest. 111, 1795–1803 (2003).
Sturm, A. C. et al. Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 72, 662–680 (2018).
Grundy, S. M. et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 73, e285–e350 (2019).
Samani, N. J. et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357, 443–453 (2007).
McPherson, R. et al. A common allele on chromosome 9 associated with coronary heart disease. Science 316, 1488–1491 (2007).
Helgadottir, A. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Tobacco and Genetics Consortium.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 42, 441–447 (2010).
Köttgen, A. et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42, 376–384 (2010).
Hellwege, J. N. et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat. Commun. 10, 3842 (2019).
Warren, H. R. et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 49, 403–415 (2017).
Ehret, G. B. et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. 48, 1171–1184 (2016).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Klarin, D. et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 50, 1514–1523 (2018).
Gibson, G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13, 135–145 (2012).
Pe’er, I., Yelensky, R., Altshuler, D. & Daly, M. J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Koyama, S. et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 52, 1169–1177 (2020).
Abraham, G. & Inouye, M. Genomic risk prediction of complex human disease and its clinical application. Curr. Opin. Genet. Dev. 33, 10–16 (2015).
Khera, A. V. et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N. Engl. J. Med. 375, 2349–2358 (2016).
Jostins, L. & Barrett, J. C. Genetic risk prediction in complex disease. Hum. Mol. Genet. 20, R182–R188 (2011).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Mak, T. S. H., Porsch, R. M., Choi, S. W., Zhou, X. & Sham, P. C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol. 41, 469–480 (2017).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Pattee, J. & Pan, W. Penalized regression and model selection methods for polygenic scores on summary statistics. PLoS Comput. Biol. 16, e1008271 (2020).
Márquez-Luna, C., Loh, P. R. & Price, A. L. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 41, 811–823 (2017).
Hu, Y. et al. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. PLoS Genet. 13, e1006836 (2017).
Hu, Y. et al. Leveraging functional annotations in genetic risk prediction for human complex diseases. PLoS Comput. Biol. 13, e1005589 (2017).
Ye, Y. et al. Interactions between enhanced polygenic risk scores and lifestyle for cardiovascular disease, diabetes, and lipid levels. Circ. Genom. Precis. Med. 14, e003128 (2021).
Márquez-Luna, C. et al. Incorporating functional priors improves polygenic prediction accuracy in UK Biobank and 23andMe data sets. Nat. Commun. 12, 6052 (2021).
Amariuta, T. et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat. Genet. 52, 1346–1354 (2020).
Wand, H. et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 591, 211–219 (2021).
Choi, S. W., Mak, T. S. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772 (2020).
Riaz, M. et al. Effect of APOE and a polygenic risk score on incident dementia and cognitive decline in a healthy older population. Aging Cell 20, e13384 (2021).
Neumann, J. T. et al. Predictive performance of a polygenic risk score for incident ischemic stroke in a healthy older population. Stroke 52, 2882–2891 (2021).
Natarajan, P. et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 135, 2091–2101 (2017).
Mars, N. et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med. 26, 549–557 (2020).
Jiang, X., Holmes, C. & McVean, G. The impact of age on genetic risk for common diseases. PLoS Genet. 17, e1009723 (2021).
Stone, N. J. et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 129 (Suppl. 2), S1–S45 (2014).
Hindy, G. et al. Genome-wide polygenic score, clinical risk factors, and long-term trajectories of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 40, 2738–2746 (2020).
Elliott, J. et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA 323, 636–645 (2020).
Mosley, J. D. et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA 323, 627–635 (2020).
Riveros-Mckay, F. et al. Integrated polygenic tool substantially enhances coronary artery disease prediction. Circ. Genom. Precis. Med. 14, e003304 (2021).
Khera, A. V. et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation 139, 1593–1602 (2019).
Homburger, J. R. et al. Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores. Genome Med. 11, 74 (2019).
Arnett, D. K. et al. 2019 ACC/AHA Guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 74, e177–e232 (2019).
Baigent, C. et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366, 1267–1278 (2005).
Mihaylova, B. et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet 380, 581–590 (2012).
Mega, J. L. et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 385, 2264–2271 (2015).
Aragam, K. G. et al. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J. Am. Coll. Cardiol. 75, 2769–2780 (2020).
Kullo, I. J. et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial). Circulation 133, 1181–1188 (2016).
Tada, H. et al. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur. Heart J. 37, 561–567 (2016).
Ripatti, P. et al. Polygenic hyperlipidemias and coronary artery disease risk. Circ. Genom. Precis. Med. 13, e002725 (2020).
CARDIoGRAMplusC4D Consortium. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Aragam, K. G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Preprint at medRxiv https://doi.org/10.1101/2021.05.24.21257377 (2021).
Karmali, K. N., Goff, D. C. Jr, Ning, H. & Lloyd-Jones, D. M. A systematic examination of the 2013 ACC/AHA pooled cohort risk assessment tool for atherosclerotic cardiovascular disease. J. Am. Coll. Cardiol. 64, 959–968 (2014).
Goldstein, B. A., Knowles, J. W., Salfati, E., Ioannidis, J. P. & Assimes, T. L. Simple, standardized incorporation of genetic risk into non-genetic risk prediction tools for complex traits: coronary heart disease as an example. Front. Genet. 5, 254 (2014).
Said, M. A., Verweij, N. & van der Harst, P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK Biobank Study. JAMA Cardiol. 3, 693–702 (2018).
Merino, J. et al. Interaction between type 2 diabetes prevention strategies and genetic determinants of coronary artery disease on cardiometabolic risk factors. Diabetes 69, 112–120 (2020).
Robinson, C. L. et al. Disclosing genetic risk for coronary heart disease: effects on perceived personal control and genetic counseling satisfaction. Clin. Genet. 89, 251–257 (2016).
Brown, S. N., Jouni, H., Marroush, T. S. & Kullo, I. J. Effect of disclosing genetic risk for coronary heart disease on information seeking and sharing: the MI-GENES Study (Myocardial Infarction Genes). Circ. Cardiovasc. Genet. 10, e001613 (2017).
Grant, R. W. et al. Personalized genetic risk counseling to motivate diabetes prevention: a randomized trial. Diabetes Care 36, 13–19 (2013).
Widén, E. et al. Communicating polygenic and non-genetic risk for atherosclerotic cardiovascular disease — an observational follow-up study. Preprint at medRxiv https://doi.org/10.1101/2020.09.18.20197137 (2020).
Zhou, X. et al. Cost-effectiveness of diabetes prevention interventions targeting high-risk individuals and whole populations: a systematic review. Diabetes Care 43, 1593–1616 (2020).
Herman, W. H. et al. The cost-effectiveness of lifestyle modification or metformin in preventing type 2 diabetes in adults with impaired glucose tolerance. Ann. Intern. Med. 142, 323–332 (2005).
Zeng, W. et al. Benefits and costs of intensive lifestyle modification programs for symptomatic coronary disease in Medicare beneficiaries. Am. Heart J. 165, 785–792 (2013).
Detrano, R. et al. Coronary calcium as a predictor of coronary events in four racial or ethnic groups. N. Engl. J. Med. 358, 1336–1345 (2008).
McClelland, R. L., Chung, H., Detrano, R., Post, W. & Kronmal, R. A. Distribution of coronary artery calcium by race, gender, and age: results from the Multi-Ethnic Study of Atherosclerosis (MESA). Circulation 113, 30–37 (2006).
Miedema, M. D. et al. Association of coronary artery calcium with long-term, cause-specific mortality among young adults. JAMA Netw. Open 2, e197440 (2019).
Paixao, A. R. et al. Coronary artery calcium improves risk classification in younger populations. JACC Cardiovasc. Imaging 8, 1285–1293 (2015).
Tota-Maharaj, R. et al. Coronary artery calcium for the prediction of mortality in young adults <45 years old and elderly adults >75 years old. Eur. Heart J. 33, 2955–2962 (2012).
Nasir, K. et al. Coronary artery calcification and family history of premature coronary heart disease: sibling history is more strongly associated than parental history. Circulation 110, 2150–2156 (2004).
Klarin, D. et al. Genetic architecture of abdominal aortic aneurysm in the Million Veteran Program. Circulation 142, 1633–1646 (2020).
Owens, D. K. et al. Screening for abdominal aortic aneurysm: US Preventive Services Task Force Recommendation Statement. JAMA 322, 2211–2218 (2019).
Besseling, J., Hovingh, G. K., Huijgen, R., Kastelein, J. J. P. & Hutten, B. A. Statins in familial hypercholesterolemia: consequences for coronary artery disease and all-cause mortality. J. Am. Coll. Cardiol. 68, 252–260 (2016).
Klarin, D. et al. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat. Genet. 51, 1574–1579 (2019).
Muse, E. D. et al. Response to polygenic risk: results of the mygenerank mobile application-based coronary artery disease study. Preprint at medRxiv https://doi.org/10.1101/2021.04.26.21256141 (2021).
Mach, F. et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur. Heart J. 41, 111–188 (2020).
Pearson, G. J. et al. 2021 Canadian Cardiovascular Society guidelines for the management of dyslipidemia for the prevention of cardiovascular disease in the adult. Can. J. Cardiol. 37, 1129–1150 (2021).
Marston, N. A. et al. Predicting benefit from evolocumab therapy in patients with atherosclerotic disease using a genetic risk score: results from the FOURIER trial. Circulation 141, 616–623 (2020).
Damask, A. et al. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation 141, 624–636 (2020).
Knowles, J. W. et al. Impact of a genetic risk score for coronary artery disease on reducing cardiovascular risk: a pilot randomized controlled study. Front. Cardiovasc. Med. 4, 53 (2017).
Moore, T. J., Zhang, H., Anderson, G. & Alexander, G. C. Estimated costs of pivotal trials for novel therapeutic agents approved by the US Food and Drug Administration, 2015-2016. JAMA Intern. Med. 178, 1451–1457 (2018).
Schwartz, G. G. et al. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N. Engl. J. Med. 379, 2097–2107 (2018).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Kent, D. M., Steyerberg, E. & van Klaveren, D. Personalized evidence based medicine: predictive approaches to heterogeneous treatment effects. BMJ 363, k4245 (2018).
Bothwell, L. E., Avorn, J., Khan, N. F. & Kesselheim, A. S. Adaptive design clinical trials: a review of the literature and ClinicalTrials.gov. BMJ Open 8, e018320 (2018).
Holmes, D. R. Jr et al. The 21st Century Cures Act and and early feasibility studies for cardiovascular devices: what have we learned, where do we need to go? JACC Cardiovasc. Interv. 11, 2220–2225 (2018).
D’Agostino, R. B. Sr. The delayed-start study design. N. Engl. J. Med. 361, 1304–1306 (2009).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04331535 (2021).
Tunis, S. R., Stryer, D. B. & Clancy, C. M. Practical clinical trials: increasing the value of clinical research for decision making in clinical and health policy. JAMA 290, 1624–1632 (2003).
Maron, D. J. et al. Initial invasive or conservative strategy for stable coronary disease. N. Engl. J. Med. 382, 1395–1407 (2020).
Al-Lamee, R. et al. Percutaneous coronary intervention in stable angina (ORBITA): a double-blind, randomised controlled trial. Lancet 391, 31–40 (2018).
Boden, W. E. et al. Optimal medical therapy with or without PCI for stable coronary disease. N. Engl. J. Med. 356, 1503–1516 (2007).
Franklin, B. A. Lessons learned from the COURAGE trial: generalizability, limitations, and implications. Prev. Cardiol. 10, 117–120 (2007).
Weintraub, W. S. et al. Effect of PCI on quality of life in patients with stable coronary disease. N. Engl. J. Med. 359, 677–687 (2008).
Levin, M. G. et al. Genomic risk stratification predicts all-cause mortality after cardiac catheterization. Circ. Genom. Precis. Med. 11, e002352 (2018).
Ford, I. & Norrie, J. Pragmatic trials. N. Engl. J. Med. 375, 454–463 (2016).
Pereira, N. L. et al. Effect of genotype-guided oral P2Y12 inhibitor selection vs conventional clopidogrel therapy on ischemic outcomes after percutaneous coronary intervention: the TAILOR-PCI randomized clinical trial. JAMA 324, 761–771 (2020).
Pereira, N. L. et al. Effect of CYP2C19 genotype on ischemic outcomes during oral P2Y12 inhibitor therapy: a meta-analysis. JACC Cardiovasc. Interv. 14, 739–750 (2021).
Executive Committee for the Asymptomatic Carotid Atherosclerosis Study. Endarterectomy for asymptomatic carotid artery stenosis. JAMA 273, 1421–1428 (1995).
Halliday, A. et al. Prevention of disabling and fatal strokes by successful carotid endarterectomy in patients without recent neurological symptoms: randomised controlled trial. Lancet 363, 1491–1502 (2004).
Brott, T. G. et al. Stenting versus endarterectomy for treatment of carotid-artery stenosis. N. Engl. J. Med. 363, 11–23 (2010).
Schermerhorn, M. L. et al. Association of transcarotid artery revascularization vs transfemoral carotid artery stenting with stroke or death among patients with carotid artery stenosis. JAMA 322, 2313–2322 (2019).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020).
Curtis, D. Polygenic risk score for schizophrenia is more strongly associated with ancestry than with schizophrenia. Psychiatr. Genet. 28, 85–89 (2018).
Schultz, L. M. et al. Stability of polygenic scores across discovery genome-wide association studies. Preprint at bioRxiv https://doi.org/10.1101/2021.06.18.449060 (2021).
Ruan, Y. et al. Improving polygenic prediction in ancestrally diverse populations. Preprint at medRxiv https://doi.org/10.1101/2020.12.27.20248738 (2021).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature 590, 290–299 (2021).
Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
All of Us Research Program Investigators. The “All of Us” research program. N. Engl. J. Med. 381, 668–676 (2019).
McCarty, C. A. et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics 4, 13 (2011).
Vujkovic, M. et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 52, 680–691 (2020).
Chen, S. F. et al. Genotype imputation and variability in polygenic risk score estimation. Genome Med. 12, 100 (2020).
Kowalski, M. H. et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 15, e1008500 (2019).
Marnetto, D. et al. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat. Commun. 11, 1628 (2020).
Lambert, S. A. et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 53, 420–425 (2021).
Carver, R. B., Castéra, J., Gericke, N., Evangelista, N. A. & El-Hani, C. N. Young adults’ belief in genetic determinism, and knowledge and attitudes towards modern genetics and genomics: the PUGGS questionnaire. PLoS ONE 12, e0169808 (2017).
Kaufman, D. J., Baker, R., Milner, L. C., Devaney, S. & Hudson, K. L. A survey of US adults’ opinions about conduct of a Nationwide Precision Medicine Initiative® cohort study of genes and environment. PLoS ONE 11, e0160461 (2016).
Author information
Authors and Affiliations
Contributions
Both authors contributed substantially to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Cardiology thanks Guillaume Paré, Ali Torkamani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
All Of Us: https://allofus.nih.gov/
eMERGE: https://emerge-network.org/
MVP: https://www.research.va.gov/mvp/
Polygenic Score Catalog: https://www.pgscatalog.org/
TOPMed: https://www.nhlbi.nih.gov/science/trans-omics-precision-medicine-topmed-program
Glossary
- Genome-wide association studies
-
(GWAS). Studies that search, in an agnostic fashion, for allelic association with a particular phenotype by genotyping tag single-nucleotide polymorphisms across the entire genome.
- Single-nucleotide polymorphism
-
(SNP). Specific substitution of a single nucleotide at a specific position in the genome.
- Imputation
-
A technique that leverages the linkage disequilibrium between genotyped and ungenotyped variants to statistically infer missing genotypes using a reference panel of genotyped individuals.
- Non-genotyped alleles
-
Single-nucleotide polymorphisms that can be inferred through statistical imputation but not directly observed on a genotype array.
- Genome-wide significance
-
A level of statistical significance required to establish association for a common variant in genome-wide association studies (P = 5 × 10−8).
- Linkage disequilibrium
-
(LD). The non-random association of alleles at two or more loci because of infrequent recombination events between them.
- Least absolute shrinkage and selection operator
-
A modelling procedure for linear regression encouraging model sparsity.
- Elastic net
-
Regularized regression method combining penalties of least absolute shrinkage and selection operator and ridge methods in a linear fashion.
- Haplotypes
-
Combinations of alleles transmitted together on a single chromosome.
- Pleiotropy
-
A single gene or variant yielding two or more apparently unrelated effects.
- Whole-genome sequencing
-
Identification of all base pairs for an individual, with subsequent mapping of contiguous reads to a reference genome sequence (for next-generation sequencing).
- Admixed populations
-
Populations in which previously diverged genetic lineages are mixed, for example, the AMR (admixed American) superpopulation in the 1,000 Genomes reference panel.
Rights and permissions
About this article
Cite this article
Klarin, D., Natarajan, P. Clinical utility of polygenic risk scores for coronary artery disease. Nat Rev Cardiol 19, 291–301 (2022). https://doi.org/10.1038/s41569-021-00638-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-021-00638-w
This article is cited by
-
Predicting the presence of coronary plaques featuring high-risk characteristics using polygenic risk scores and targeted proteomics in patients with suspected coronary artery disease
Genome Medicine (2024)
-
Genetic variation across and within individuals
Nature Reviews Genetics (2024)
-
Integration of polygenic and gut metagenomic risk prediction for common diseases
Nature Aging (2024)
-
The Role of Genetics in Advancing Cardiometabolic Drug Development
Current Atherosclerosis Reports (2024)
-
What Causes Premature Coronary Artery Disease?
Current Atherosclerosis Reports (2024)