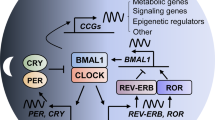
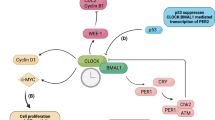
Abstract
Circadian rhythms govern a large array of physiological and metabolic functions. Perturbations of the daily cycle have been linked to elevated risk of developing cancer as well as poor prognosis in patients with cancer. Also, expression of core clock genes or proteins is remarkably attenuated particularly in tumours of a higher stage or that are more aggressive, possibly linking the circadian clock to cellular differentiation. Emerging evidence indicates that metabolic control by the circadian clock underpins specific hallmarks of cancer metabolism. Indeed, to support cell proliferation and biomass production, the clock may direct metabolic processes of cancer cells in concert with non-clock transcription factors to control how nutrients and metabolites are utilized in a time-specific manner. We hypothesize that the metabolic switch between differentiation or stemness of cancer may be coupled to the molecular clockwork. Moreover, circadian rhythms of host organisms appear to dictate tumour growth and proliferation. This Review outlines recent discoveries of the interplay between circadian rhythms, proliferative metabolism and cancer, highlighting potential opportunities in the development of future therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fu, L. & Lee, C. C. The circadian clock: pacemaker and tumour suppressor. Nat. Rev. Cancer 3, 350–361 (2003).
Masri, S., Kinouchi, K. & Sassone-Corsi, P. Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56 (2015).
Masri, S. & Sassone-Corsi, P. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 24, 1795–1803 (2018).
Hirota, T. et al. Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012).
Solt, L. A. et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012).
Gill, S. & Panda, S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 22, 789–798 (2015).
Greco, C. M. & Sassone-Corsi, P. Circadian blueprint of metabolic pathways in the brain. Nat. Rev. Neurosci. 20, 71–82 (2019).
Gaucher, J., Montellier, E. & Sassone-Corsi, P. Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol. 28, 368–379 (2018).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998).
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000).
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010).
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002).
Schibler, U. & Sassone-Corsi, P. A web of circadian pacemakers. Cell 111, 919–922 (2002). This review captures the original view of how circadian clocks are organized in a complex network that may be essential for their fundamental properties of timekeeping and entrainment to environmental signals.
Asher, G. & Sassone-Corsi, P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015).
Damiola, F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). This study illustrates that peripheral clocks are entrained by timed food intake and highlights the role of food as a zeitgeber.
Stokkan, K. A., Yamazaki, S., Tei, H., Sakaki, Y. & Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001).
Eckel-Mahan, K. L. et al. Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). This study uncovers that the circadian machinery can be reprogrammed transcriptionally and metabolically by food intake.
Gaucher, J. et al. Distinct metabolic adaptation of liver circadian pathways to acute and chronic patterns of alcohol intake. Proc. Natl Acad. Sci. USA 116, 25250–25259 (2019).
Tognini, P. et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 26, 523–538.e5 (2017).
Guan, D. et al. Diet-induced circadian enhancer remodeling synchronizes opposing hepatic lipid metabolic processes. Cell 174, 831–842.e12 (2018).
Kohsaka, A. et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007).
Dyar, K. A. et al. Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018). This study uses high-throughput metabolomics to build the first atlas of circadian metabolism and reveals the importance of temporal metabolic coordination across different tissues.
Koronowski, K. B. et al. Defining the independence of the liver circadian clock. Cell 177, 1448–1462.e14 (2019).
Welz, P. S. et al. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 177, 1436–1447.e12 (2019). Together with Koronowski et al. (2019), this paper establishes the degree of autonomy of peripheral circadian clocks in otherwise clock-deficient mice.
Ueda, H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005).
Ukai-Tadenuma, M., Kasukawa, T. & Ueda, H. R. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 10, 1154–1163 (2008).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). This study identifies the parallel between circadian transcriptional regulation and temporal patterns of genome-wide occupancy of core circadian clock regulators.
Ripperger, J. A. & Schibler, U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 (2006).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012).
Ueda, H. R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002).
Zhang, Y. et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015).
Fang, B. et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014). This study investigates circadian enhancer RNAs and demonstrates the commitment of diverse TFs to temporal gene regulation.
Kume, K. et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 (1999).
Ukai-Tadenuma, M. et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144, 268–281 (2011).
van der Horst, G. T. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999).
Shigeyoshi, Y. et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053 (1997).
Crosio, C., Cermakian, N., Allis, C. D. & Sassone-Corsi, P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 3, 1241–1247 (2000).
Ginty, D. D. et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241 (1993).
Sato, M., Murakami, M., Node, K., Matsumura, R. & Akashi, M. The role of the endocrine system in feeding-induced tissue-specific circadian entrainment. Cell Rep. 8, 393–401 (2014).
Balsalobre, A. et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000).
Zhang, E. E. et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 (2010).
Adamovich, Y., Ladeuix, B., Golik, M., Koeners, M. P. & Asher, G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 25, 93–101 (2017).
Buhr, E. D., Yoo, S. H. & Takahashi, J. S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330, 379–385 (2010).
Husse, J., Leliavski, A., Tsang, A. H., Oster, H. & Eichele, G. The light–dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J. 28, 4950–4960 (2014).
Ishida, A. et al. Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab. 2, 297–307 (2005). This paper illustrates a hypothalamus–pituitary–adrenal axis-independent, photic induction of glucocorticoid release from the adrenal gland.
Kolbe, I., Leinweber, B., Brandenburger, M. & Oster, H. Circadian clock network desynchrony promotes weight gain and alters glucose homeostasis in mice. Mol. Metab. 30, 140–151 (2019).
Terazono, H. et al. Adrenergic regulation of clock gene expression in mouse liver. Proc. Natl Acad. Sci. USA 100, 6795–6800 (2003).
Balsalobre, A., Marcacci, L. & Schibler, U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 10, 1291–1294 (2000).
Robles, M. S., Humphrey, S. J. & Mann, M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 25, 118–127 (2017).
Lamia, K. A. et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 (2009).
Lipton, J. O. et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161, 1138–1151 (2015).
Sahar, S., Zocchi, L., Kinoshita, C., Borrelli, E. & Sassone-Corsi, P. Regulation of BMAL1 protein stability and circadian function by GSK3β-mediated phosphorylation. PLoS ONE 5, e8561 (2010).
Menzies, K. J., Zhang, H., Katsyuba, E. & Auwerx, J. Protein acetylation in metabolism—metabolites and cofactors. Nat. Rev. Endocrinol. 12, 43–60 (2016).
Pietrocola, F., Galluzzi, L., Bravo-San Pedro, J. M., Madeo, F. & Kroemer, G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 (2015).
Asher, G. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008).
Hirayama, J. et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 (2007).
Nakahata, Y. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008).
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M. & Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK–SIRT1. Science 324, 654–657 (2009).
Ramsey, K. M. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 (2009). Together with Nakahata et al. (2009), this paper uncovers the link between the core circadian clock and the cyclic production of NAD+ through a metabolic feedback loop.
Fustin, J. M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013).
Hirano, A., Braas, D., Fu, Y. H. & Ptacek, L. J. FAD regulates CRYPTOCHROME protein stability and circadian clock in mice. Cell Rep. 19, 255–266 (2017).
Kaasik, K. et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302 (2013).
Li, M. D. et al. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303–310 (2013).
Cao, Q. et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 69, 7619–7625 (2009).
de Assis, L. V. M. et al. Non-metastatic cutaneous melanoma induces chronodisruption in central and peripheral circadian clocks. Int. J. Mol. Sci. 19, 1065 (2018).
Oh, E. Y. et al. Circadian transcription profile of mouse breast cancer under light–dark and dark–dark conditions. Cancer Genomics Proteomics 7, 311–322 (2010).
Anafi, R. C., Francey, L. J., Hogenesch, J. B. & Kim, J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc. Natl Acad. Sci. USA 114, 5312–5317 (2017).
Shilts, J., Chen, G. & Hughey, J. J. Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ 6, e4327 (2018).
Ye, Y. et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 6, 314–328.e2 (2018).
Abreu, M., Basti, A., Genov, N., Mazzoccoli, G. & Relogio, A. The reciprocal interplay between TNFα and the circadian clock impacts on cell proliferation and migration in Hodgkin lymphoma cells. Sci. Rep. 8, 11474 (2018).
Rida, P., Syed, M. I. & Aneja, R. Time will tell: circadian clock dysregulation in triple negative breast cancer. Front. Biosci. 11, 178–192 (2019).
de The, H. Differentiation therapy revisited. Nat. Rev. Cancer 18, 117–127 (2018).
Dierickx, P., Van Laake, L. W. & Geijsen, N. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 19, 18–28 (2018).
Sladek, M., Jindrakova, Z., Bendova, Z. & Sumova, A. Postnatal ontogenesis of the circadian clock within the rat liver. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R1224–R1229 (2007).
Umemura, Y. et al. Transcriptional program of Kpna2/Importin-α2 regulates cellular differentiation-coupled circadian clock development in mammalian cells. Proc. Natl Acad. Sci. USA 111, E5039–E5048 (2014).
Umemura, Y. et al. Involvement of posttranscriptional regulation of Clock in the emergence of circadian clock oscillation during mouse development. Proc. Natl Acad. Sci. USA 114, E7479–E7488 (2017).
Yagita, K. et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc. Natl Acad. Sci. USA 107, 3846–3851 (2010).
de Assis, L. V. M. et al. Expression of the circadian clock gene BMAL1 positively correlates with antitumor immunity and patient survival in metastatic melanoma. Front. Oncol. 8, 185 (2018).
Li, W. et al. Decreased circadian component Bmal1 predicts tumor progression and poor prognosis in human pancreatic ductal adenocarcinoma. Biochem. Biophys. Res. Commun. 472, 156–162 (2016).
Mazzoccoli, G. et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol. Int. 28, 841–851 (2011).
Zeng, Z. L. et al. Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. 20, 1042–1052 (2014).
Fekry, B. et al. Incompatibility of the circadian protein BMAL1 and HNF4α in hepatocellular carcinoma. Nat. Commun. 9, 4349 (2018).
Taniguchi, H. et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 69, 8447–8454 (2009).
Yeh, C. M. et al. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int. J. Oncol. 45, 2101–2107 (2014).
Zhang, S. et al. Circadian clock components RORα and Bmal1 mediate the anti-proliferative effect of MLN4924 in osteosarcoma cells. Oncotarget 7, 66087–66099 (2016).
Zeng, Z. L. et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J. Biochem. 148, 319–326 (2010).
Ohashi, M. et al. Disruption of circadian clockwork in in vivo reprogramming-induced mouse kidney tumors. Genes Cell 23, 60–69 (2018).
Bu, Y. et al. A PERK–miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat. Cell Biol. 20, 104–115 (2018).
Yamada, Y., Haga, H. & Yamada, Y. Concise review: dedifferentiation meets cancer development: proof of concept for epigenetic cancer. Stem Cell Transl Med. 3, 1182–1187 (2014).
Dai, H. et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res. Treat. 127, 531–540 (2011).
Alhopuro, P. et al. Mutations in the circadian gene CLOCK in colorectal cancer. Mol. Cancer Res. 8, 952–960 (2010).
Puram, R. V. et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell 165, 303–316 (2016).
Gery, S. et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 22, 375–382 (2006).
Oshima, T. et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol. Rep. 25, 1439–1446 (2011).
Xia, H. C. et al. Deregulated expression of the Per1 and Per2 in human gliomas. Can. J. Neurol. Sci. 37, 365–370 (2010).
Zhao, H. et al. Prognostic relevance of Period1 (Per1) and Period2 (Per2) expression in human gastric cancer. Int. J. Clin. Exp. Pathol. 7, 619–630 (2014).
Broadberry, E. et al. Disrupted circadian clocks and altered tissue mechanics in primary human breast tumours. Breast Cancer Res. 20, 125 (2018).
Gery, S. et al. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood 106, 2827–2836 (2005).
Xiang, R. et al. Circadian clock gene Per2 downregulation in nonsmall cell lung cancer is associated with tumour progression and metastasis. Oncol. Rep. 40, 3040–3048 (2018).
Gery, S., Virk, R. K., Chumakov, K., Yu, A. & Koeffler, H. P. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 26, 7916–7920 (2007).
Hua, H. et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 97, 589–596 (2006).
Miyazaki, K., Wakabayashi, M., Hara, Y. & Ishida, N. Tumor growth suppression in vivo by overexpression of the circadian component, PER2. Genes Cells 15, 351–358 (2010).
Oda, A. et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 29, 1201–1209 (2009).
Cadenas, C. et al. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle 13, 3282–3291 (2014).
Fu, L., Pelicano, H., Liu, J., Huang, P. & Lee, C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 (2002). This study demonstrates that loss of the Per2 gene leads to mice prone to cancer.
Gu, X. et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 19, 397–405 (2012).
Wood, P. A. et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol. Cancer Res. 6, 1786–1793 (2008).
Hua, H. et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 14, 815–818 (2007).
Qin, T. et al. Effect of Period 2 on the proliferation, apoptosis and migration of osteosarcoma cells, and the corresponding mechanisms. Oncol. Lett. 16, 2668–2674 (2018).
Yang, X. et al. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol. Int. 26, 1323–1339 (2009).
Yang, X. et al. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res. Treat. 117, 423–431 (2009).
Hwang-Verslues, W. W. et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl Acad. Sci. USA 110, 12331–12336 (2013).
Grimaldi, B. et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 12, 509–520 (2010).
Gotoh, T. et al. Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor Period 2. Proc. Natl Acad. Sci. USA 113, 13516–13521 (2016).
Travnickova-Bendova, Z., Cermakian, N., Reppert, S. M. & Sassone-Corsi, P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl Acad. Sci. USA 99, 7728–7733 (2002).
Yang, W. S. & Stockwell, B. R. Inhibition of casein kinase 1-ε induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 9, R92 (2008).
Liu, J. et al. Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci. Signal. 11, eaau0715 (2018).
Lahav, G. et al. Dynamics of the p53–Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147–150 (2004).
Dyar, K. A. et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29–41 (2014).
Yu, H. et al. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. PLoS ONE 8, e61679 (2013).
Mannic, T. et al. Circadian clock characteristics are altered in human thyroid malignant nodules. J. Clin. Endocrinol. Metab. 98, 4446–4456 (2013).
Ozturk, N., Lee, J. H., Gaddameedhi, S. & Sancar, A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc. Natl Acad. Sci. USA 106, 2841–2846 (2009).
Lamia, K. A. et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011).
Chun, S. K. et al. A synthetic cryptochrome inhibitor induces anti-proliferative effects and increases chemosensitivity in human breast cancer cells. Biochem. Biophys. Res. Commun. 467, 441–446 (2015).
Kriebs, A. et al. Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl Acad. Sci. USA 114, 8776–8781 (2017).
Busino, L. et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 (2007).
Godinho, S. I. et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007).
Siepka, S. M. et al. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023 (2007).
Huber, A. L. et al. CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016). This study identifies a possible mechanism through which the circadian clock might inhibit MYC.
Kettner, N. M. et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016).
Lee, S., Donehower, L. A., Herron, A. J., Moore, D. D. & Fu, L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS ONE 5, e10995 (2010).
Chen, S. T. et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26, 1241–1246 (2005).
Zhu, Y. et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol. Int. 28, 852–861 (2011).
Rao, R. C. & Dou, Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346 (2015).
Katada, S. & Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421 (2010).
Valekunja, U. K. et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl Acad. Sci. USA 110, 1554–1559 (2013).
Miki, T. et al. PML regulates PER2 nuclear localization and circadian function. EMBO J. 31, 1427–1439 (2012).
Vander Heiden, M. G. & DeBerardinis, R. J. Understanding the Intersections between metabolism and cancer biology. Cell 168, 657–669 (2017). This review article discusses limiting metabolic pathways in cancer cell proliferation.
Tohyama, S. et al. Glutamine oxidation is indispensable for survival of human pluripotent stem cells. Cell Metab. 23, 663–674 (2016).
Tohyama, S., Tanosaki, S., Someya, S., Fujita, J. & Fukuda, K. Manipulation of pluripotent stem cell metabolism for clinical application. Curr. Stem Cell Rep. 3, 28–34 (2017).
Peek, C. B. et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417 (2013). This study demonstrates that loss of Bmal1 induces a metabolic switch from lipid oxidation to glycolysis.
Doi, R., Oishi, K. & Ishida, N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J. Biol. Chem. 285, 22114–22121 (2010).
Marcheva, B. et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 (2010).
Walton, Z. E. et al. Acid suspends the circadian clock in hypoxia through inhibition of mTOR. Cell 174, 72–87.e32 (2018). This study shows that the acid–base balance influences the circadian clock function.
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Imai, S. Armstrong, C. M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 795–800 (2000).
Elhanati, S. et al. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 4, 905–912 (2013).
Masri, S. et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 (2014).
Ponugoti, B. et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 285, 33959–33970 (2010).
Currie, E., Schulze, A., Zechner, R., Walther, T. C. & Farese, R. V. Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 (2013).
Svensson, R. U. et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 (2016).
Bugge, A. et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 (2012).
Sulli, G. et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553, 351–355 (2018).
Dierickx, P. et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc. Natl Acad. Sci. USA 116, 12147–12152 (2019).
Dang, C. V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 3, a014217 (2013).
Gu, W. et al. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell 19, 476–490 (2016).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Altman, B. J. et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 (2015). This study provides a molecular underpinning by which MYC inhibits oscillation of the circadian clock.
Repouskou, A. & Prombona, A. c-MYC targets the central oscillator gene Per1 and is regulated by the circadian clock at the post-transcriptional level. Biochim. Biophys. Acta 1859, 541–552 (2016).
Shostak, A. et al. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat. Commun. 7, 11807 (2016).
Hsieh, A. L. et al. Misregulation of Drosophila Myc disrupts circadian behavior and metabolism. Cell Rep. 29, 1778–1788.e4 (2019).
Papagiannakopoulos, T. et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 24, 324–331 (2016). This study reveals that Per2 loss of function promotes metabolic reprogramming in mouse lung cancer cells.
Keith, B., Johnson, R. S. & Simon, M. C. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12, 9–22 (2011).
Hu, C. J., Wang, L. Y., Chodosh, L. A., Keith, B. & Simon, M. C. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell Biol. 23, 9361–9374 (2003).
Iyer, N. V. et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 (1998).
Rankin, E. B. & Giaccia, A. J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 15, 678–685 (2008).
Ryan, H. E., Lo, J. & Johnson, R. S. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17, 3005–3015 (1998).
Peek, C. B. et al. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 25, 86–92 (2017).
Wu, Y. et al. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 25, 73–85 (2017).
Kobayashi, M. et al. A circadian clock gene, PER2, activates HIF-1 as an effector molecule for recruitment of HIF-1α to promoter regions of its downstream genes. FEBS J. 284, 3804–3816 (2017).
Sato, S. et al. Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell Metab. 30, 92–110.e4 (2019).
Manella, G. et al. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc. Natl Acad. Sci. USA 117, 779–786 (2020).
Almendros, I. et al. Intermittent hypoxia increases melanoma metastasis to the lung in a mouse model of sleep apnea. Respir. Physiol. Neurobiol. 186, 303–307 (2013).
Karoor, V. et al. Alveolar hypoxia promotes murine lung tumor growth through a VEGFR-2/EGFR-dependent mechanism. Cancer Prev. Res. 5, 1061–1071 (2012).
Bersten, D. C., Sullivan, A. E., Peet, D. J. & Whitelaw, M. L. bHLH–PAS proteins in cancer. Nat. Rev. Cancer 13, 827–841 (2013).
King, A., Selak, M. A. & Gottlieb, E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25, 4675–4682 (2006).
Tabebi, M., Soderkvist, P. & Jensen, L. D. Hypoxia signaling and circadian disruption in and by pheochromocytoma. Front. Endocrinol. 9, 612 (2018).
Zhikrivetskaya, S. O. et al. Molecular markers of paragangliomas/pheochromocytomas. Oncotarget 8, 25756–25782 (2017).
Opitz, C. A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Kuiper, R. P. et al. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 12, 1661–1669 (2003).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009).
Sidhar, S. K. et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum. Mol. Genet. 5, 1333–1338 (1996).
Weterman, M. A., Wilbrink, M. & Geurts van Kessel, A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proc. Natl Acad. Sci. USA 93, 15294–15298 (1996).
Pastore, N. et al. Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. EMBO J. 38, e101347 (2019).
Michael, A. K. et al. Cancer/testis antigen PASD1 silences the circadian clock. Mol. Cell 58, 743–754 (2015).
Hansen, J. Light at night, shiftwork, and breast cancer risk. J. Natl Cancer Inst. 93, 1513–1515 (2001).
Knutsson, A. et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand. J. Work Environ. Health 39, 170–177 (2013).
Megdal, S. P., Kroenke, C. H., Laden, F., Pukkala, E. & Schernhammer, E. S. Night work and breast cancer risk: a systematic review and meta-analysis. Eur. J. Cancer 41, 2023–2032 (2005).
Rafnsson, V., Tulinius, H., Jonasson, J. G. & Hrafnkelsson, J. Risk of breast cancer in female flight attendants: a population-based study (Iceland). Cancer Causes Control 12, 95–101 (2001).
Schernhammer, E. S. et al. Rotating night shifts and risk of breast cancer in women participating in the Nurses’ Health Study. J. Natl Cancer Inst. 93, 1563–1568 (2001).
Srour, B. et al. Circadian nutritional behaviours and cancer risk: new insights from the NutriNet-Sante prospective cohort study: disclaimers. Int. J. Cancer 143, 2369–2379 (2018).
Viswanathan, A. N. & Schernhammer, E. S. Circulating melatonin and the risk of breast and endometrial cancer in women. Cancer Lett. 281, 1–7 (2009).
Viswanathan, A. N., Hankinson, S. E. & Schernhammer, E. S. Night shift work and the risk of endometrial cancer. Cancer Res. 67, 10618–10622 (2007).
Conlon, M., Lightfoot, N. & Kreiger, N. Rotating shift work and risk of prostate cancer. Epidemiology 18, 182–183 (2007).
Kloog, I., Haim, A., Stevens, R. G. & Portnov, B. A. Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiol. Int. 26, 108–125 (2009).
Kubo, T. et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am. J. Epidemiol. 164, 549–555 (2006).
Schernhammer, E. S. et al. Night-shift work and risk of colorectal cancer in the Nurses’ Health Study. J. Natl Cancer Inst. 95, 825–828 (2003).
Lahti, T. A., Partonen, T., Kyyronen, P., Kauppinen, T. & Pukkala, E. Night-time work predisposes to non-Hodgkin lymphoma. Int. J. Cancer 123, 2148–2151 (2008).
Basen-Engquist, K. & Chang, M. Obesity and cancer risk: recent review and evidence. Curr. Oncol. Rep. 13, 71–76 (2011).
Hastings, M. H., Reddy, A. B. & Maywood, E. S. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 (2003).
Filipski, E. et al. Disruption of circadian coordination accelerates malignant growth in mice. Pathol. Biol. 51, 216–219 (2003).
Filipski, E. et al. Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl Cancer Inst. 97, 507–517 (2005).
van den Heiligenberg, S. et al. The tumor promoting effect of constant light exposure on diethylnitrosamine-induced hepatocarcinogenesis in rats. Life Sci. 64, 2523–2534 (1999).
Van Dycke, K. C. et al. Chronically alternating light cycles increase breast cancer risk in mice. Curr. Biol. 25, 1932–1937 (2015).
Blask, D. E. et al. Light exposure at night disrupts host/cancer circadian regulatory dynamics: impact on the Warburg effect, lipid signaling and tumor growth prevention. PLoS ONE 9, e102776 (2014).
Filipski, E., Li, X. M. & Levi, F. Disruption of circadian coordination and malignant growth. Cancer Causes Control 17, 509–514 (2006).
Dauchy, R. T., Blask, D. E., Sauer, L. A., Brainard, G. C. & Krause, J. A. Dim light during darkness stimulates tumor progression by enhancing tumor fatty acid uptake and metabolism. Cancer Lett. 144, 131–136 (1999).
Mhatre, M. C., Shah, P. N. & Juneja, H. S. Effect of varying photoperiods on mammary morphology, DNA synthesis, and hormone profile in female rats. J. Natl Cancer Inst. 72, 1411–1416 (1984).
Proietti, S., Cucina, A., Minini, M. & Bizzarri, M. Melatonin, mitochondria, and the cancer cell. Cell. Mol. Life Sci. 74, 4015–4025 (2017).
Pevet, P. & Challet, E. Melatonin: both master clock output and internal time-giver in the circadian clocks network. J. Physiol. Paris 105, 170–182 (2011).
Talib, W. H. Melatonin and cancer hallmarks. Molecules 23, 518 (2018).
Li, Y. et al. Melatonin for the prevention and treatment of cancer. Oncotarget 8, 39896–39921 (2017).
Li, J. C. & Xu, F. Influences of light–dark shifting on the immune system, tumor growth and life span of rats, mice and fruit flies as well as on the counteraction of melatonin. Biol. Signals 6, 77–89 (1997).
Shah, P. N., Mhatre, M. C. & Kothari, L. S. Effect of melatonin on mammary carcinogenesis in intact and pinealectomized rats in varying photoperiods. Cancer Res. 44, 3403–3407 (1984).
Reddy, A. B. et al. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45, 1478–1488 (2007).
So, A. Y., Bernal, T. U., Pillsbury, M. L., Yamamoto, K. R. & Feldman, B. J. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl Acad. Sci. USA 106, 17582–17587 (2009).
Kiessling, S., Sollars, P. J. & Pickard, G. E. Light stimulates the mouse adrenal through a retinohypothalamic pathway independent of an effect on the clock in the suprachiasmatic nucleus. PLoS ONE 9, e92959 (2014).
Oster, H. et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 4, 163–173 (2006).
Son, G. H. et al. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc. Natl Acad. Sci. USA 105, 20970–20975 (2008).
Charmandari, E. et al. Peripheral CLOCK regulates target-tissue glucocorticoid receptor transcriptional activity in a circadian fashion in man. PLoS ONE 6, e25612 (2011).
Kino, T., Charmandari, E. & Chrousos, G. P. Glucocorticoid receptor: implications for rheumatic diseases. Clin. Exp. Rheumatol. 29, S32–S41 (2011).
Moreno-Smith, M., Lutgendorf, S. K. & Sood, A. K. Impact of stress on cancer metastasis. Future Oncol. 6, 1863–1881 (2010).
Yang, H. et al. Stress–glucocorticoid–TSC22D3 axis compromises therapy-induced antitumor immunity. Nat. Med. 25, 1428–1441 (2019).
Shimba, A. et al. Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity 48, 286–298.e6 (2018).
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer–immunity cycle. Immunity 39, 1–10 (2013).
Suzuki, S. et al. Circadian rhythm of leucocytes and lymphocytes subsets and its possible correlation with the function of the autonomic nervous system. Clin. Exp. Immunol. 110, 500–508 (1997).
Maestroni, G. J. et al. Neural and endogenous catecholamines in the bone marrow. Circadian association of norepinephrine with hematopoiesis? Exp. Hematol. 26, 1172–1177 (1998).
Okamoto, S., Ibaraki, K., Hayashi, S. & Saito, M. Ventromedial hypothalamus suppresses splenic lymphocyte activity through sympathetic innervation. Brain Res. 739, 308–313 (1996).
Logan, R. W. et al. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. J. Immunol. 188, 2583–2591 (2012).
Creed, S. J. et al. β2-Adrenoceptor signaling regulates invadopodia formation to enhance tumor cell invasion. Breast Cancer Res. 17, 145 (2015).
Magnon, C. et al. Autonomic nerve development contributes to prostate cancer progression. Science 341, 1236361 (2013).
Sloan, E. K. et al. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 70, 7042–7052 (2010).
Mormont, M. C. et al. Marker rhythms of circadian system function: a study of patients with metastatic colorectal cancer and good performance status. Chronobiol. Int. 19, 141–155 (2002).
Mormont, M. C. & Waterhouse, J. Contribution of the rest–activity circadian rhythm to quality of life in cancer patients. Chronobiol. Int. 19, 313–323 (2002).
Innominato, P. F. et al. Circadian rhythm in rest and activity: a biological correlate of quality of life and a predictor of survival in patients with metastatic colorectal cancer. Cancer Res. 69, 4700–4707 (2009).
Sephton, S. E., Sapolsky, R. M., Kraemer, H. C. & Spiegel, D. Diurnal cortisol rhythm as a predictor of breast cancer survival. J. Natl Cancer Inst. 92, 994–1000 (2000).
Hojo, H. et al. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget 8, 34128–34140 (2017).
Masri, S. et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 165, 896–909 (2016). This study reveals that lung tumours influence host circadian metabolism and illustrates the importance of temporal communications between the host and the tumour.
Flint, T. R. et al. Tumor-induced IL-6 reprograms host metabolism to suppress anti-tumor immunity. Cell Metab. 24, 672–684 (2016).
Ocana, M. C., Martinez-Poveda, B., Quesada, A. R. & Medina, M. A. Metabolism within the tumor microenvironment and its implication on cancer progression: an ongoing therapeutic target. Med. Res. Rev. 39, 70–113 (2019).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371.e9 (2017).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Borniger, J. C. et al. A role for hypocretin/orexin in metabolic and sleep abnormalities in a mouse model of non-metastatic breast cancer. Cell Metab. 28, 118–129 e115 (2018). This study identifies neuroendocrine pathways through which breast cancer modulates sleep and glucose homeostasis in mice.
Honma, K., Honma, S., Kohsaka, M. & Fukuda, N. Seasonal variation in the human circadian rhythm: dissociation between sleep and temperature rhythm. Am. J. Physiol. 262, R885–R891 (1992).
Noya, S. B. et al. The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science 366, eaav2642 (2019).
Hakim, F. et al. Fragmented sleep accelerates tumor growth and progression through recruitment of tumor-associated macrophages and TLR4 signaling. Cancer Res. 74, 1329–1337 (2014).
Liang, J. A. et al. Non-apnea sleep disorders will increase subsequent liver cancer risk—a nationwide population-based cohort study. Sleep Med. 13, 869–874 (2012).
Li, X. M., Vincenti, M. & Levi, F. Pharmacological effects of vinorelbine on body temperature and locomotor activity circadian rhythms in mice. Chronobiol. Int. 19, 43–55 (2002).
Lyall, M. J. et al. Diurnal profile of interstitial glucose following dexamethasone prophylaxis for chemotherapy treatment of gynaecological cancer. Diabet. Med. 35, 1508–1514 (2018).
Palesh, O. et al. Prevalence, putative mechanisms, and current management of sleep problems during chemotherapy for cancer. Nat. Sci. Sleep 4, 151–162 (2012).
Wu, M. W., Li, X. M., Xian, L. J. & Levi, F. Effects of meal timing on tumor progression in mice. Life Sci. 75, 1181–1193 (2004).
Li, X. M. et al. Cancer inhibition through circadian reprogramming of tumor transcriptome with meal timing. Cancer Res. 70, 3351–3360 (2010).
Kinouchi, K. et al. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 25, 3299–3314.e6 (2018).
Nencioni, A., Caffa, I., Cortellino, S. & Longo, V. D. Fasting and cancer: molecular mechanisms and clinical application. Nat. Rev. Cancer 18, 707–719 (2018).
Sato, S. et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e11 (2017).
Yoshino, J., Mills, K. F., Yoon, M. J. & Imai, S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 (2011).
Gui, D. Y. et al. Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity to metformin. Cell Metab. 24, 716–727 (2016).
Yang, G. et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl Med. 8, 324ra316 (2016).
Antoch, M. P. et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle 7, 1197–1204 (2008).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003).
Xu, C. X., Wang, C., Krager, S. L., Bottum, K. M. & Tischkau, S. A. Aryl hydrocarbon receptor activation attenuates Per1 gene induction and influences circadian clock resetting. Toxicol. Sci. 132, 368–378 (2013).
Tischkau, S. A., Jaeger, C. D. & Krager, S. L. Circadian clock disruption in the mouse ovary in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Lett. 201, 116–122 (2011).
Murray, I. A., Patterson, A. D. & Perdew, G. H. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer 14, 801–814 (2014).
Hatori, M. et al. Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Proc. Natl Acad. Sci. USA 108, 4864–4869 (2011).
Gilardi, F. et al. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 10, e1004155 (2014).
Le Martelot, G. et al. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7, e1000181 (2009).
Chen, M. et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 50, 206–218 (2018).
Kauffman, E. C. et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat. Rev. Urol. 11, 465–475 (2014).
Hochegger, H., Takeda, S. & Hunt, T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat. Rev. Mol. Cell Biol. 9, 910–916 (2008).
Grechez-Cassiau, A., Rayet, B., Guillaumond, F., Teboul, M. & Delaunay, F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283, 4535–4542 (2008).
Kowalska, E. et al. NONO couples the circadian clock to the cell cycle. Proc. Natl Acad. Sci. USA 110, 1592–1599 (2013).
Matsuo, T. et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 (2003).
Plikus, M. V. et al. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc. Natl Acad. Sci. USA 110, E2106–E2115 (2013).
Geyfman, M. et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl Acad. Sci. USA 109, 11758–11763 (2012).
Feillet, C. et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc. Natl Acad. Sci. USA 111, 9828–9833 (2014).
Nagoshi, E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004).
Bieler, J. et al. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 10, 739 (2014).
Acknowledgements
The authors thank S. Masri for insight and advice. They thank all members of the Sassone-Corsi laboratory for discussions and support. K.K. was supported by a JSPS fellowship. The authors’ research is supported by the National Institutes of Health (NIH), a Novo Nordisk Challenge Grant and INSERM (France).
Author information
Authors and Affiliations
Contributions
P.S.-C. and K.K. researched data for the article, made substantial contributions to discussion of content and wrote, reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Kenichiro Kinouchi dedicates this article to the memory of Paolo Sassone-Corsi, for his outstanding contributions and scientific advancements in the fields of circadian rhythms, epigenetics and metabolism, and expresses the deepest appreciation for his mentorship and generosity.
Peer review information
Nature Reviews Cancer thanks J. Borniger, C. Dang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Diurnal feeding–fasting behaviour
-
The daily behaviour of feeding and fasting under day–night cycles.
- Hypothalamic suprachiasmatic nucleus
-
A group of oscillator neurons that govern circadian rhythms of physiology and behaviour.
- Autonomic nervous system
-
A network of peripheral nerves that govern fundamental organ functions, often unconsciously.
- Sympathetic nervous system
-
A stimulatory branch of the autonomic nervous system that is activated in fight or flight response.
- Lipogenesis
-
A reductive metabolic process of synthesizing lipids by gaining electrons frequently activated in proliferating cells.
- Lysosomal dispersion
-
The peripheral redistribution of normally perinuclear lysosomes.
- Cancer/testis antigen
-
A class of antigenic proteins present on some human cancers but not on adult normal tissues except for testes.
- Polysynaptic pathways
-
Neural pathways comprising multiple neural connections in which signals are transmitted.
- Ventromedial hypothalamus
-
A specialized hypothalamic nucleus that controls appetite and thermogenesis.
- Ketogenesis
-
A metabolic process that produces ketone bodies from fatty acids or amino acids.
- Hypocretin/orexin neurons
-
A group of neurons in the lateral hypothalamus that regulate wakefulness and appetite.
Rights and permissions
About this article
Cite this article
Kinouchi, K., Sassone-Corsi, P. Metabolic rivalry: circadian homeostasis and tumorigenesis. Nat Rev Cancer 20, 645–661 (2020). https://doi.org/10.1038/s41568-020-0291-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-020-0291-9
This article is cited by
-
Neural function of Bmal1: an overview
Cell & Bioscience (2023)
-
Six-hour time-restricted feeding inhibits lung cancer progression and reshapes circadian metabolism
BMC Medicine (2023)
-
Nucleus-exported CLOCK acetylates PRPS to promote de novo nucleotide synthesis and liver tumour growth
Nature Cell Biology (2023)
-
Cellular rejuvenation: molecular mechanisms and potential therapeutic interventions for diseases
Signal Transduction and Targeted Therapy (2023)
-
Circadian disruption: from mouse models to molecular mechanisms and cancer therapeutic targets
Cancer and Metastasis Reviews (2023)