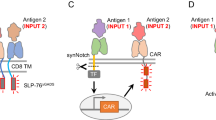
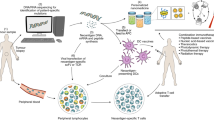
Abstract
Engineered immune-cell-based cancer therapies have demonstrated robust efficacy in B cell malignancies, but challenges such as the lack of ideal targetable tumour antigens, tumour-mediated immunosuppression and severe toxicity still hinder their therapeutic efficacy and broad applicability. Synthetic biology can be used to overcome these challenges and create more robust, effective adaptive therapies that enable the specific targeting of cancer cells while sparing healthy cells. In this Progress article, we review recently developed gene circuit therapies for cancer using immune cells, nucleic acids and bacteria as chassis. We conclude by discussing outstanding challenges and future directions for realizing these gene circuit therapies in the clinic.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kitada, T., DiAndreth, B., Teague, B. & Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Auslander, S. & Fussenegger, M. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 31, 155–168 (2013).
Kittleson, J. T., Wu, G. C. & Anderson, J. C. Successes and failures in modular genetic engineering. Curr. Opin. Chem. Biol. 16, 329–336 (2012).
Porter, D. L., Levine, B. L., Kalos, M., Bagg, A. & June, C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011).
Grupp, S. A. et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 368, 1509–1518 (2013).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Neelapu, S. S. et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017).
Schuster, S. J. et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N. Engl. J. Med. 377, 2545–2554 (2017).
Giavridis, T. et al. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 24, 731–738 (2018).
Lim, W. A. & June, C. H. The principles of engineering immune cells to treat cancer. Cell 168, 724–740 (2017).
Chakravarti, D., Cho, J. H., Weinberg, B. H., Wong, N. M. & Wong, W. W. Synthetic biology approaches in cancer immunotherapy, genetic network engineering, and genome editing. Integr. Biol. 8, 504–517 (2016).
Roybal, K. T. & Lim, W. A. Synthetic immunology: hacking immune cells to expand their therapeutic capabilities. Annu. Rev. Immunol. 35, 229–253 (2017).
Klebanoff, C. A., Rosenberg, S. A. & Restifo, N. P. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat. Med. 22, 26–36 (2016).
Tasian, S. K. & Gardner, R. A. CD19-redirected chimeric antigen receptor-modified T cells: a promising immunotherapy for children and adults with B cell acute lymphoblastic leukemia (ALL). Ther. Adv. Hematol. 6, 228–241 (2015).
Morgan, R. A. et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 18, 843–851 (2010).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791 (2016).
Roybal, K. T. et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016).
Kloss, C. C., Condomines, M., Cartellieri, M., Bachmann, M. & Sadelain, M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 31, 71–75 (2013).
Fedorov, V. D., Themeli, M. & Sadelain, M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl Med. 5, 215ra172 (2013).
Gardner, R. A. et al. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood 129, 3322–3331 (2017).
Lee, D. W. et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015).
Hegde, M. et al. Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T cells in glioblastoma. Mol. Ther. 21, 2087–2101 (2013).
Grada, Z. et al. TanCAR: a novel bispecific chimeric antigen receptor for cancer immunotherapy. Mol. Ther. Nucleic Acids 2, e105 (2013).
Zah, E., Lin, M. Y., Silva-Benedict, A., Jensen, M. C. & Chen, Y. Y. T. Cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol. Res. 4, 498–508 (2016).
Qin, H., Haso, W., Nguyen, S. M. & Fry, T. J. Preclinical development of bispecific chimeric antigen receptor targeting both CD19 and CD22. Blood 126, 4427–4427 (2015).
Cho, J. H., Collins, J. J. & Wong, W. W. Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell 173, 1426–1438 (2018).
Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014).
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016).
Strønen, E. et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341 (2016).
Ho, P., Ede, C. & Chen, Y. Y. Modularly constructed synthetic granzyme B molecule enables interrogation of intracellular proteases for targeted cytotoxicity. ACS Synth. Biol. 6, 1484–1495 (2017).
Norelli, M. et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 24, 739–748 (2018).
Di Stasi, A. et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 365, 1673–1683 (2011).
Wu, C. Y., Roybal, K. T., Puchner, E. M., Onuffer, J. & Lim, W. A. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015).
Pan, Y. et al. Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc. Natl Acad. Sci. USA 115, 992–997 (2018).
Park, J. S. et al. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc. Natl Acad. Sci. USA 111, 5896–5901 (2014).
Rabinovich, G. A., Gabrilovich, D. & Sotomayor, E. M. Immunosuppressive strategies that are mediated by tumor cells. Annu. Rev. Immunol. 25, 267–296 (2007).
Yeku, O. O. & Brentjens, R. J. Armored CAR T cells: utilizing cytokines and pro-inflammatory ligands to enhance CAR T cell anti-tumour efficacy. Biochem. Soc. Trans. 44, 412–418 (2016).
Zhang, L. et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res. 21, 2278–2288 (2015).
Roybal, K. T. et al. Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell 167, 419–432.e16 (2016).
Chang, Z. L. et al. Rewiring T cell responses to soluble factors with chimeric antigen receptors. Nat. Chem. Biol. 14, 317–324 (2018).
Prosser, M. E., Brown, C. E., Shami, A. F., Forman, S. J. & Jensen, M. C. Tumor PD-L1 co-stimulates primary human CD8(+) cytotoxic T cells modified to express a PD1:CD28 chimeric receptor. Mol. Immunol. 51, 263–272 (2012).
Ankri, C., Shamalov, K., Horovitz-Fried, M., Mauer, S. & Cohen, C. J. Human T cells engineered to express a programmed death 1/28 costimulatory retargeting molecule display enhanced antitumor activity. J. Immunol. 191, 4121–4129 (2013).
Anderson, K. G., Stromnes, I. M. & Greenberg, P. D. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell 31, 311–325 (2017).
Kojima, R., Scheller, L. & Fussenegger, M. Nonimmune cells equipped with T cell-receptor-like signaling for cancer cell ablation. Nat. Chem. Biol. 14, 42–49 (2018).
Nissim, L. & Bar-Ziv, R. H. A tunable dual-promoter integrator for targeting of cancer cells. Mol. Syst. Biol. 6, 444 (2010).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011).
Wroblewska, L. et al. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 33, 839–841 (2015).
Baudrimont, A. et al. Multiplexed gene control reveals rapid mRNA turnover. Sci. Adv. 3, e1700006 (2017).
Mircetic, J., Dietrich, A., Paszkowski-Rogacz, M., Krause, M. & Buchholz, F. Development of a genetic sensor that eliminates p53 deficient cells. Nat. Commun. 8, 1463 (2017).
Liu, Y. et al. Directing cellular information flow via CRISPR signal conductors. Nat. Methods 13, 938–944 (2016).
Liu, Y., Li, J., Chen, Z., Huang, W. & Cai, Z. Synthesizing artificial devices that redirect cellular information at will. eLife 7, e31936 (2018).
Dastor, M. et al. A workflow for in vivo evaluation of candidate inputs and outputs for cell classifier gene circuits. ACS Synth. Biol. 7, 474–489 (2018).
Nissim, L. et al. Synthetic RNA-based immunomodulatory gene circuits for cancer immunotherapy. Cell 171, 1138–1150 (2017).
Nakazawa, Y. et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J. Immunother. 32, 826–836 (2009).
Ivics, Z. & Izsvak, Z. The expanding universe of transposon technologies for gene and cell engineering. Mob. DNA 1, 25 (2010).
Eyquem, J. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017).
MacLeod, D. T. et al. Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther. 25, 949–961 (2017).
Qasim, W. et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl Med. 9, eaaj2013 (2017).
Ren, J. et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266 (2017).
Fang, F., Xiao, W. & Tian, Z. NK cell-based immunotherapy for cancer. Semin. Immunol. 31, 37–54 (2017).
Krueger, T. E. G., Thorek, D. L. J., Denmeade, S. R., Isaacs, J. T. & Brennen, W. N. Concise review: mesenchymal stem cell-based drug delivery: the good, the bad, the ugly, and the promise. Stem Cells Transl Med. 7, 651–663 (2018).
Ando, M. & Nakauchi, H. ‘Off-the-shelf’ immunotherapy with iPSC-derived rejuvenated cytotoxic T lymphocytes. Exp. Hematol. 47, 2–12 (2017).
Ibraheem, D., Elaissari, A. & Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 459, 70–83 (2014).
Lundstrom, K. Viral vectors in gene therapy. Diseases 6, E42 (2018).
Lawler, S. E., Speranza, M. C., Cho, C. F. & Chiocca, E. A. Oncolytic viruses in cancer treatment: a review. JAMA Oncol. 3, 841–849 (2017).
Shi, B. et al. Challenges in DNA delivery and recent advances in multifunctional polymeric DNA delivery systems. Biomacromolecules 18, 2231–2246 (2017).
Yin, H. et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014).
Wagner, T. E. et al. Small-molecule-based regulation of RNA-delivered circuits in mammalian cells. Nat. Chem. Biol. 14, 1043–1050 (2018).
Schreiber, J., Arter, M., Lapique, N., Haefliger, B. & Benenson, Y. Model-guided combinatorial optimization of complex synthetic gene networks. Mol. Syst. Biol. 12, 899 (2016).
Kramer, M. G., Masner, M., Ferreira, F. A. & Hoffman, R. M. Bacterial therapy of cancer: promises, limitations, and insights for future directions. Front. Microbiol. 9, 16 (2018).
Anderson, J. C., Clarke, E. J., Arkin, A. P. & Voigt, C. A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 355, 619–627 (2006).
Swofford, C. A., Van Dessel, N. & Forbes, N. S. Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc. Natl Acad. Sci. USA 112, 3457–3462 (2015).
Din, M. O. et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016).
Huh, J. H., Kittleson, J. T., Arkin, A. P. & Anderson, J. C. Modular design of a synthetic payload delivery device. ACS Synth. Biol. 2, 418–424 (2013).
Shi, L., Yu, B., Cai, C. H. & Huang, J. D. Angiogenic inhibitors delivered by the type III secretion system of tumor-targeting Salmonella typhimurium safely shrink tumors in mice. AMB Express 6, 56 (2016).
Xu, X. et al. Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res. 74, 6260–6270 (2014).
West, K. A. et al. Abstract 2920: Metabolic modulation of the tumor microenvironment using Synthetic Biotic™ Medicines. Cancer Res. 78, 2920 (2018).
Siska, P. J. & Rathmell, J. C. T cell metabolic fitness in antitumor immunity. Trends Immunol. 36, 257–264 (2015).
Leventhal, D. et al. Activation of innate and adaptive immunity via combinatorial immunotherapy using Synthetic Biotic™ Medicines. Cancer Res. 78, LB-131 (2018).
Chen, Q., Sun, L. & Chen, Z. J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142 (2016).
Grushkin, D. The new drug circuit. Nat. Med. 18, 1452 (2012).
Sadelain, M., Brentjens, R. & Riviere, I. The basic principles of chimeric antigen receptor design. Cancer Discov. 3, 388–398 (2013).
Acknowledgements
T.K.L. is supported by the Department of Defense (W81XWH-16-1-0565 and W81XWH-18-1-0513) and the Koch Institute for Integrative Cancer Research at Massachusetts Institute of Technology Bridge Project. M.-R.W. is supported by the Department of Defense (W81XWH-16-1-0452).
Author information
Authors and Affiliations
Contributions
M.-R.W. and B.J. contributed equally. All authors discussed the contents and wrote the article.
Corresponding author
Ethics declarations
Competing interests
M.-R.W. and T.K.L. have filed patent applications on part of work discussed in this article. T.K.L. is a co-founder of BiomX, Corvium, Eligo Biosciences, Engine Biosciences, Senti Biosciences, Synlogic and Tango Therapeutics. T.K.L. also holds financial interests in AmpliPhi, IndieBio and Nest.Bio. B.J. declares no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, MR., Jusiak, B. & Lu, T. Engineering advanced cancer therapies with synthetic biology. Nat Rev Cancer 19, 187–195 (2019). https://doi.org/10.1038/s41568-019-0121-0
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-019-0121-0