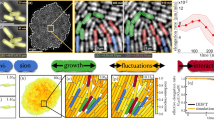
Abstract
To maximize their fitness, cells must be able to respond effectively to stresses. This demands making trade-offs between processes that conserve resources to promote survival, and processes that use resources to promote growth and division. Understanding the nature of these trade-offs and the physics underlying them remains an outstanding challenge. Here we combine single-cell experiments and theoretical modelling to propose a mechanism for antibiotic adaptation through mechanical feedback between cell growth and morphology. Under long-term exposure to sublethal doses of ribosome-targeting antibiotics, we find that Caulobacter crescentus cells can recover their pre-stimulus growth rates and undergo dramatic changes in cell shape. Upon antibiotic removal, cells recover their original forms over multiple generations. These phenomena are explained by a physical theory of bacterial growth, which demonstrates that an increase in cell width and curvature promotes faster growth under protein synthesis inhibition. Shape changes thereby make bacteria more adaptive to surviving antibiotics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data are provided with this paper. All other data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Code availability
Custom computer codes that were used in this paper are available from the corresponding authors upon reasonable request.
References
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Jun, S., Si, F., Pugatch, R. & Scott, M. Fundamental principles in bacterial physiology—history, recent progress, and the future with focus on cell size control: a review. Rep. Prog. Phys. 81, 056601 (2018).
Willis, L. & Huang, K. C. Sizing up the bacterial cell cycle. Nat. Rev. Microbiol. 15, 606–620 (2017).
Young, K. D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006).
Yang, D. C., Blair, K. M. & Salama, N. R. Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80, 187–203 (2016).
Woldemeskel, S. A. & Goley, E. D. Shapeshifting to survive: shape determination and regulation in Caulobacter crescentus. Trends Microbiol. 25, 673–687 (2017).
Deforet, M., van Ditmarsch, D. & Xavier, J. B. Cell-size homeostasis and the incremental rule in a bacterial pathogen. Biophys. J. 109, 521–528 (2015).
Lock, R. L. & Harry, E. J. Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7, 324–338 (2008).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Kohanski, M. A., DePristo, M. A. & Collins, J. J. Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37, 311–320 (2010).
Zhang, Q. et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333, 1764–1767 (2011).
Toprak, E. et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 44, 101–105 (2012).
Deris, J. B. et al. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342, 1237435 (2013).
Greulich, P., Scott, M., Evans, M. R. & Allen, R. J. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 11, 796 (2015).
Nonejuie, P., Burkart, M., Pogliano, K. & Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl Acad. Sci. USA 110, 16169–16174 (2013).
Yao, Z., Kahne, D. & Kishony, R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol. Cell 48, 705–712 (2012).
Si, F. et al. Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr. Biol. 27, 1278–1287 (2017).
Harris, L. K. & Theriot, J. A. Relative rates of surface and volume synthesis set bacterial cell size. Cell 165, 1479–1492 (2016).
Harris, L. K. & Theriot, J. A. Surface area to volume ratio: a natural variable for bacterial morphogenesis. Trends Microbiol. 26, 815–832 (2018).
Wright, C. S. et al. Intergenerational continuity of cell shape dynamics in Caulobacter crescentus. Sci. Rep. 5, 9155 (2015).
Banerjee, S. et al. Biphasic growth dynamics control cell division in Caulobacter crescentus. Nat. Microbiol. 2, 17116 (2017).
Lin, Y., Crosson, S. & Scherer, N. F. Single-gene tuning of Caulobacter cell cycle period and noise, swarming motility, and surface adhesion. Mol. Syst. Biol. 6, 445 (2010).
Iyer-Biswas, S. et al. Scaling laws governing stochastic growth and division of single bacterial cells. Proc. Natl Acad. Sci. USA 111, 15912–15917 (2014).
Sliusarenko, O., Cabeen, M. T., Wolgemuth, C. W., Jacobs-Wagner, C. & Emonet, T. Processivity of peptidoglycan synthesis provides a built-in mechanism for the robustness of straight-rod cell morphology. Proc. Natl Acad. Sci. USA 107, 10086–10091 (2010).
Ursell, T. S. et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl Acad. Sci. USA 111, E1025–E1034 (2014).
Shi, H. et al. Deep phenotypic mapping of bacterial cytoskeletal mutants reveals physiological robustness to cell size. Curr. Biol. 27, 3419–3429 (2017).
Wong, F. et al. Mechanical strain sensing implicated in cell shape recovery in Escherichia coli. Nat. Microbiol. 2, 17115 (2017).
Ojkic, N., Serbanescu, D. & Banerjee, S. Surface-to-volume scaling and aspect ratio preservation in rod-shaped bacteria. eLife 8, e47033 (2019).
Tu, Y. & Rappel, W.-J. Adaptation in living systems. Annu. Rev. Condens. Matter Phys. 9, 183–205 (2018).
Jiang, H. & Sun, S. X. Morphology, growth, and size limit of bacterial cells. Phys. Rev. Lett. 105, 028101 (2010).
Banerjee, S., Scherer, N. F. & Dinner, A. R. Shape dynamics of growing cell walls. Soft Matter 12, 3442–3450 (2016).
Garner, E. C. et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 (2012).
Pinette, M. F. & Koch, A. L. Turgor pressure responses of a gram-negative bacterium to antibiotic treatment, measured by collapse of gas vesicles. J. Bacteriol. 170, 1129–1136 (1988).
Hocking, J. et al. Osmolality-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J. Bacteriol. 194, 3116–3127 (2012).
Koshland, D. E.Jr., Goldbeter, A. & Stock, J. B. Amplification and adaptation in regulatory and sensory systems. Science 217, 220–225 (1982).
Barkai, N. & Leibler, S. Robustness in simple biochemical networks. Nature 387, 913–917 (1997).
Lan, G., Sartori, P., Neumann, S., Sourjik, V. & Tu, Y. The energy–speed–accuracy trade-off in sensory adaptation. Nat. Phys. 8, 422–428 2012).
Rojas, E. R., Huang, K. C. & Theriot, J. A. Homeostatic cell growth is accomplished mechanically through membrane tension inhibition of cell-wall synthesis. Cell Syst. 5, 578–590 (2017).
Campos, M. et al. A constant size extension drives bacterial cell size homeostasis. Cell 159, 1433–1446 (2014).
Heinrich, K., Leslie, D. J., Morlock, M., Bertilsson, S. & Jonas, K. Molecular basis and ecological relevance of Caulobacter cell filamentation in freshwater habitats. mBio 10, e01557-19 (2019).
Harris, L. K., Dye, N. A. & Theriot, J. A. A Caulobacter MreB mutant with irregular cell shape exhibits compensatory widening to maintain a preferred surface area to volume ratio. Mol. Microbiol. 94, 988–1005 (2014).
Schaechter, M., Maaløe, O. & Kjeldgaard, N. O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19, 592–606 (1958).
Basan, M. et al. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 11, 836 (2015).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
Deng, Y., Sun, M. & Shaevitz, J. W. Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett. 107, 158101 (2011).
Acknowledgements
We thank L. Harris for providing additional data for C. crescentus under chloramphenicol treatment. We thank C. Wright and S. Iyer-Biswas for experimental data from C. crescentus single-cell measurements and C. Nikas for assistance with data analysis. S.B. acknowledges support from the Engineering and Physical Sciences Research Council of the United Kingdom (grant no. EP/R029822/1), Royal Society University Research Fellowship (URF/R1/180187), and Royal Society Fellows Enhancement Award (grant no. RGF/EA/181044). A.R.D. and N.F.S. acknowledge funding from the National Science Foundation Physics of Living Systems Program (NSF PHY-1305542) and from the National Science Foundation Materials Research Science and Engineering Center at the University of Chicago (NSF DMR-1420709 and NSF DMR-2011854). A.R.D. also acknowledges support from National Science Foundation award MCB-1953402.
Author information
Authors and Affiliations
Contributions
S.B., N.F.S. and A.R.D. designed the study. A.R.D. and N.F.S. designed the experiments. S.B. developed the theory. K.L. performed experiments. S.B., N.O. and R.S. performed model simulations. S.B., K.L., N.O. and R.S. analysed the data. S.B., N.F.S. and A.R.D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Physics thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
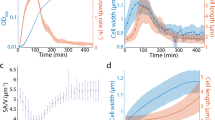
Extended Data Fig. 1 Cell shape, size control and growth dynamics during antibiotic adaptation, shown in real time.
a, Cell elongation rate, κ, as a function of absolute time for CHL concentrations: 0.1 μg/ml (blue, Number of cells n=40, Total number of generations g=941) and 0.5 μg/ml (red, n=135, g=986). Error bars indicate ± 1 SEM. b, Interdivision time, τ, as a function of absolute time. c, Cell length at birth, L(0), as a function of absolute time. d, Correlation between cell length at division, L(τ), and cell length at birth, L(0), is best described by a mixer model: L(τ)=1.1 L(0) +0.75 μm. e, Spatiotemporally averaged cell diameter (width), w, as a function of absolute time. f, Cell-cycle averaged cell curvature, R−1, as a function of absolute time.
Extended Data Fig. 2 Dynamics of cell shape and growth rate in response to mechano-chemical perturbations.
Model predictions for the response of (a) growth rate κ, (b) curvature R−1, and (c) width w, to perturbations in parameters: {ε,kc} (blue), ε (green), {ε,kL} (purple), {ε, kc, kL} (red), and {ε, kc,P} (black). Perturbation to a particular parameter μ is of the form μ → μ/(1+ϕ) for t > ta, where μ ∈ {ε, kc, kL,P}. The translation inhibitors used experimentally for Figure 1 likely affect parameters ε and kc.
Extended Data Fig. 3 Effect of turgor pressure on cellular response to chloramphenicol.
Intergenerational dynamics of (a) growth rate κ, (b) average cell width w, (c) average curvature R−1 and (d) length at birth L(0) in response to a step pulse of 0.1 μg/ml CHL applied at t=450 min for three different cases: turgor pressure remains unchanged (blue solid circles), turgor pressure is reduced by 25% by CHL (red solid circles), and turgor pressure is increased by 25% by CHL (green data points). Turgor pressure reduction leads to a decrease in cell diameter, inconsistent with experimental data. Moderate increase in turgor pressure is consistent with experimental data.
Extended Data Fig. 4 Cell width modulation alone is not sufficient to achieve growth rate adaptation.
Intergenerational dynamics of (a) growth rate κ, (b) average cell width w, and (c) average curvature R−1 in response to a step pulse of 0.1 μg/ml CHL applied at t=450 min for two different cases: Cell curvature is variable and adapts to CHL-induced growth inhibition (blue data points) and curvature is constant and not affected by CHL (red data points). In the absence of curvature modulation, adaptive response is much weaker.
Extended Data Fig. 5 Coupling the physical model for bacterial growth with a biochemical model for chloramphenicol-ribosome interactions.
a, Schematic of the biochemical pathway of ribosome-CHL interaction. CHL with extracellular concentration aex enters the cell with net flux proportional to (Pin aex- Poutain)A/V, where Pin and Pout are the inward and outward permeabilities of the cell envelope. CHL binds to ribosomes at a rate kon and unbinds with a rate koff. Growth rate is linearly proportional to the fraction of unbound ribosomes. Ribosomes upregulate their synthesis when a fraction of them are bound to CHL. Model A: No mechanical feedback between cell shape and growth rate. Model B: Cell elongation promotes an increase in surface stress σ which in turn inhibits growth rate. b-f, Intergenerational dynamics of (b) growth rate κ, (c) intracellular CHL concentration ain, (d) concentration of active ribosomes, (e) average cell width w, and (f) average curvature R−1 in response to a step pulse of 0.1 μg/ml CHL applied at t=450 min for Model A (blue) and Model B (red). g, Cell shape evolution simulated using Model B (time progression: left-to-right and top-to-bottom), shows antibiotic dilution. Color coding indicates the intracellular concentration of CHL.
Extended Data Fig. 6 Speed-accuracy trade-off in antibiotic adaptation.
a, Adaptation error (post-stimulus recovery error %) for κ, R, w and L as a function of antibiotic stress, ϕ. b, Rate of adaptation (in units of generation-1) as a function of ϕ. c, Trade-off between adaptation speed (defined as the rate of recovery) and adaptation accuracy (defined as 100-Error%).
Extended Data Fig. 7 Quantitative comparisons between single-cell simulations and experimental data for pulsatory chloramphenicol dose.
a-b, Cell growth rate κ (a) and interdivision time τ (b) upon application of a step dose of 0.1 μg/ml chloramphenicol. Blue: experimental data, Orange: Simulation data with ϕ=0.8. c-d, Cell growth rate (c) and interdivision time (d) for a pulsatile antibiotic dose of 0.5 μg/ml. Blue: experimental data, Orange: Simulation data with ϕ=3.0. Error bars indicate ± 1 standard deviation.
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2, Figs. 1–3 and Tables 1–3.
Source data
Source Data Fig. 1
Experimental time course data for Caulobacter cell shape and growth rate under chloramphenicol treatment.
Source Data Fig. 2
Correlation data between cell growth rate and curvature.
Source Data Fig. 3
Simulated data for cell shape and growth rate under chloramphenicol treatment.
Source Data Fig. 4
Experimental and simulated time course data for Caulobacter cell shape and growth rate under pulsatile chloramphenicol stress.
Source Data Extended Data Fig. 1
Experimental data for Caulobacter cell shape and growth rate under chloramphenicol treatment, plotted in real time.
Rights and permissions
About this article
Cite this article
Banerjee, S., Lo, K., Ojkic, N. et al. Mechanical feedback promotes bacterial adaptation to antibiotics. Nat. Phys. 17, 403–409 (2021). https://doi.org/10.1038/s41567-020-01079-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-020-01079-x
This article is cited by
-
Morphologic design of nanostructures for enhanced antimicrobial activity
Journal of Nanobiotechnology (2022)
-
Microbial adaptation to different environmental conditions: molecular perspective of evolved genetic and cellular systems
Archives of Microbiology (2022)