Abstract
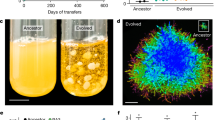
The evolution of multicellularity set the stage for sustained increases in organismal complexity1,2,3,4,5. However, a fundamental aspect of this transition remains largely unknown: how do simple clusters of cells evolve increased size when confronted by forces capable of breaking intracellular bonds? Here we show that multicellular snowflake yeast clusters6,7,8 fracture due to crowding-induced mechanical stress. Over seven weeks (~291 generations) of daily selection for large size, snowflake clusters evolve to increase their radius 1.7-fold by reducing the accumulation of internal stress. During this period, cells within the clusters evolve to be more elongated, concomitant with a decrease in the cellular volume fraction of the clusters. The associated increase in free space reduces the internal stress caused by cellular growth, thus delaying fracture and increasing cluster size. This work demonstrates how readily natural selection finds simple, physical solutions to spatial constraints that limit the evolution of group size—a fundamental step in the evolution of multicellularity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Szathmary, E. & Smith, J. M. The major evolutionary transitions. Nature 374, 227–232 (1995).
Kirk, D. L. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 27, 299–310 (2005).
Willensdorfer, M. Organism size promotes the evolution of specialized cells in multicellular digital organisms. J. Evol. Biol. 21, 104–110 (2008).
Bonner, J. T. The origins of multicellularity. Integr. Biol. 1, 27–36 (1998).
Fairclough, S. R., Dayel, M. J. & King, N. Multicellular development in a choanoflagellate. Curr. Biol. 20, R875–R876 (2010).
Ratcliff, W. C., Denison, R. F., Borrello, M. & Travisano, M. Experimental evolution of multicellularity. Proc. Natl Acad. Sci. USA 109, 1595–1600 (2012).
Ratcliff, W. C., Fankhauser, J. D., Rogers, D. W., Greig, D. & Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 6, 6102 (2015).
Ratcliff, W. C., Pentz, J. T. & Travisano, M. Tempo and mode of multicellular adaptation in experimentally evolved Saccharomyces cerevisiae. Evolution 67, 1573–1581 (2013).
Grosberg, R. K. & Strathmann, R. R. The evolution of multicellularity: A minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654 (2007).
Herron, M. D., Hackett, J. D., Aylward, F. O. & Michod, R. E. Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl Acad. Sci. USA 106, 3254–3258 (2009).
Koschwanez, J. H., Foster, K. R. & Murray, A. W. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife 2, e00367 (2013).
Hanschen, E. R. et al. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 7, 11370 (2016).
Kirkegaard, J. B., Bouillant, A., Marron, A. O., Leptos, K. C. & Goldstein, R. E. Aerotaxis in the closest relatives of animals. eLife 5, e18109 (2016).
Anderson, D. P. et al. Evolution of an ancient protein function involved in organized multicellularity in animals. eLife 5, e10147 (2016).
Withers, P. Ja. B. & Bhadeshia, H. K. D. H. Residual stress part 1 – measurement techniques. Mater. Sci. Technol. 17, 355–365 (2001).
Ahmad, M. R., Nakajima, M., Kojima, S., Homma, M. & Fukuda, T. The effects of cell sizes, environmental conditions, and growth phases on the strength of individual W303 yeast cells inside ESEM. IEEE Trans. Nanobiosci. 7, 185–193 (2008).
Delarue, M. et al. Self-driven Jjamming in growing microbial populations. Nat. Phys. 12, 762–766 (2016).
O’Hern, C. S., Silbert, L. E., Liu, A. J. & Nagel, S. R. Jamming at zero temperature and zero applied stress: the epitome of disorder. Phys. Rev. E 68, 011306 (2003).
Libby, E., Ratcliff, W., Travisano, M. & Kerr, B. Geometry shapes evolution of early multicellularity. PLoS Comput. Biol. 10, e1003803 (2014).
Sheu, Y. J., Barral, Y. & Snyder, M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247 (2000).
Donev, A. et al. Improving the density of jammed disordered packings using ellipsoids. Science 303, 990–993 (2004).
Smith, W. P. et al. Cell morphology drives spatial patterning in microbial communities. Proc. Natl Acad. Sci. USA 114, E280–E286 (2017).
Lenski, R. E. & Travisano, M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814 (1994).
Gerrish, P. J. & Lenski, R. E. The fate of competing beneficial mutations in an asexual population. Genetica 102–103, 127–144 (1998).
Kessin, R. H., Gundersen, G. G., Zaydfudim, V. & Grimson, M. How cellular slime molds evade nematodes. Proc. Natl Acad. Sci. USA 93, 4857–4861 (1996).
Boraas, M. E., Seale, D. B. & Boxhorn, J. E. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol. Ecol. 12, 153–164 (1998).
Smukalla, S. et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 726–737 (2008).
Pfeiffer, T. & Bonhoeffer, S. An evolutionary scenario for the transition to undifferentiated multicellularity. Proc. Natl Acad. Sci. USA 100, 1095–1098 (2003).
Ornes, S. Core concept: how nonequilibrium thermodynamics speaks to the mystery of life. Proc. Natl Acad. Sci. USA 114, 423–424 (2017).
Buchanan, M. Simple yet successful. Nat. Phys. 13, 720 (2017).
Transtrum, M. K. et al. Perspective: sloppiness and emergent theories in physics, biology, and beyond. J. Chem. Phys. 143, 010901 (2015).
Mattingly, H. H., Transtrum, M. K., Abbott, M. C. & Machta, B. B. Rational ignorance: simpler models learn more information from finite data. Preprint at https://arxiv.org/abs/1705.01166 (2017).
Acknowledgements
This work was supported by NASA Exobiology grant no. NNX15AR33G to W.C.R., NSF grant no. IOS-1656549 to W.C.R. and P.J.Y., and a Packard Foundation Fellowship for W.C.R. We would like to thank J. Weitz and D. Yanni for helpful comments.
Author information
Authors and Affiliations
Contributions
S.J., W.C.R. and P.J.Y. planned this research. S.J., J.T.P. and C.G.B. performed all experiments. S.J. and C.G.B. analysed the AFM and microscopy data. E.C.G. wrote the geometric simulation; E.C.G., S.J. and P.J.Y. analysed the simulation data. S.J. and P.J.Y. wrote the first draft of the paper; all authors contributed to revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information
Supplementary Figures 1–11, Supplementary Notes and Supplementary References
Rights and permissions
About this article
Cite this article
Jacobeen, S., Pentz, J.T., Graba, E.C. et al. Cellular packing, mechanical stress and the evolution of multicellularity. Nature Phys 14, 286–290 (2018). https://doi.org/10.1038/s41567-017-0002-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-017-0002-y
This article is cited by
-
Emergence and maintenance of stable coexistence during a long-term multicellular evolution experiment
Nature Ecology & Evolution (2024)
-
De novo evolution of macroscopic multicellularity
Nature (2023)
-
Bacteria evolve macroscopic multicellularity by the genetic assimilation of phenotypically plastic cell clustering
Nature Communications (2023)
-
Drug-dependent growth curve reshaping reveals mechanisms of antifungal resistance in Saccharomyces cerevisiae
Communications Biology (2022)
-
Oxygen suppression of macroscopic multicellularity
Nature Communications (2021)