Abstract
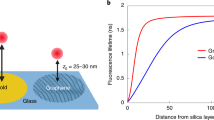
Single-molecule fluorescence imaging has become an indispensable tool for almost all fields of research, from fundamental physics to the life sciences. Among its most important applications is single-molecule localization super-resolution microscopy (SMLM) (for example, photoactivated localization microscopy (PALM)1, stochastic optical reconstruction microscopy (STORM)2, fluorescent PALM (fPALM)3, direct STORM (dSTORM)4 and point accumulation for imaging in nanoscale topography (PAINT)5), which uses the fact that the centre position of a single molecule’s image can be determined with much higher accuracy than the size of that image itself. However, a big challenge of SMLM is to achieve super-resolution along the third dimension as well. Recently, metal-induced energy transfer (MIET) was introduced to axially localize fluorescent emitters6,7,8,9. This exploits the energy transfer from an excited fluorophore to plasmons in a thin metal film. Here, we show that by using graphene as the ‘metal’ layer, one can increase the localization accuracy of MIET by nearly tenfold. We demonstrate this by axially localizing single emitters and by measuring the thickness of lipid bilayers with ångström accuracy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding authors upon reasonable request.
Code availability
All Matlab routines and codes used for data analysis of this study are available from the corresponding authors upon request.
References
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Hess, S. T., Girirajan, T. P. & Mason, M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. 47, 6172–6176 (2008).
Sharonov, A. & Hochstrasser, R. M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl Acad. Sci. USA 103, 18911–18916 (2006).
Chizhik, A. I., Rother, J., Gregor, I., Janshoff, A. & Enderlein, J. Metal-induced energy transfer for live cell nanoscopy. Nat. Photon. 8, 124–127 (2014).
Isbaner, S. et al. Axial colocalization of single molecules with nanometer accuracy using metal-induced energy transfer. Nano Lett. 18, 2616–2622 (2018).
Karedla, N. et al. Single-molecule metal-induced energy transfer (smMIET): resolving nanometer distances at the single-molecule level. ChemPhysChem 15, 705–711 (2014).
Karedla, N. et al. Three-dimensional single-molecule localization with nanometer accuracy using metal-induced energy transfer (MIET) imaging. J. Chem. Phys. 148, 204201 (2018).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Juette, M. F. et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Backlund, M. P. et al. Simultaneous, accurate measurement of the 3D position and orientation of single molecules. Proc. Natl Acad. Sci. USA 109, 19087–19092 (2012).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl Acad. Sci. USA 106, 3125–3130 (2009).
Aquino, D. et al. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat. Methods 8, 353–359 (2011).
Bourg, N. et al. Direct optical nanoscopy with axially localized detection. Nat. Photon. 9, 587–593 (2015).
Bon, P. et al. Self-interference 3D super-resolution microscopy for deep tissue investigations. Nat. Methods 15, 449–454 (2018).
Novoselov, K. S. et al. Electric field effect in atomically thin carbon films. Science 306, 666–669 (2004).
Wallace, P. R. The band theory of graphite. Phys. Rev. 71, 622 (1947).
Weber, J., Calado, V. & Van De Sanden, M. Optical constants of graphene measured by spectroscopic ellipsometry. Appl. Phys. Lett. 97, 091904 (2010).
Chance, R., Prock, A. & Silbey, R. Molecular fluorescence and energy transfer near interfaces. Adv. Chem Phys. 37, 1–65 (1978).
Enderlein, J. Single-molecule fluorescence near a metal layer. Chem. Phys. 247, 1–9 (1999).
Chizhik, A. I. et al. Electrodynamic coupling of electric dipole emitters to a fluctuating mode density within a nanocavity. Phys. Rev. Lett. 108, 163002 (2012).
Chizhik, A. I., Gregor, I., Ernst, B. & Enderlein, J. Nanocavity-based determination of absolute values of photoluminescence quantum yields. ChemPhysChem 14, 505–513 (2013).
Böhmer, M. & Enderlein, J. Orientation imaging of single molecules by wide-field epifluorescence microscopy. J. Opt. Soc. Am. B 20, 554–559 (2003).
Patra, D., Gregor, I. & Enderlein, J. Image analysis of defocused single-molecule images for three-dimensional molecule orientation studies. J. Phys. Chem. A 108, 6836 (2004).
Chizhik, A. I. et al. Probing the radiative transition of single molecules with a tunable microresonator. Nano Lett. 11, 1700–1703 (2011).
Bagatolli, L. A. To see or not to see: lateral organization of biological membranes and fluorescence microscopy. Biochim. Biophys. Acta 1758, 1541–1556 (2006).
Schneider, F., Ruhlandt, D., Gregor, I., Enderlein, J. & Chizhik, A. I. Quantum yield measurements of fluorophores in lipid bilayers using a plasmonic nanocavity. J. Phys. Chem. Lett. 8, 1472–1475 (2017).
Kučerka, N., Nieh, M.-P. & Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta 1808, 2761–2771 (2011).
Attwood, S. J., Choi, Y. & Leonenko, Z. Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy. Int. J. Mol. Sci. 14, 3514–3539 (2013).
Tristram-Nagle, S., Petrache, H. I. & Nagle, J. F. Structure and interactions of fully hydrated dioleoylphosphatidylcholine bilayers. Biophys. J. 75, 917–925 (1998).
Chizhik, A. M. et al. Three-dimensional reconstruction of nuclear envelope architecture using dual-color metal-induced energy transfer imaging. ACS Nano 11, 11839–11846 (2017).
Baronsky, T. et al. Cell–substrate dynamics of the epithelial-to-mesenchymal transition. Nano Lett. 17, 3320–3326 (2017).
Simoncelli, S., Makarova, M., Wardley, W. & Owen, D. M. Toward an axial nanoscale ruler for fluorescence microscopy. ACS Nano 11, 11762–11767 (2017).
Chizhik, A. M. et al. Dual-color metal-induced and Förster resonance energy transfer for cell nanoscopy. Mol. Biol. Cell 29, 846–851 (2018).
Balzarotti, F. et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2016).
Acknowledgements
We are grateful to the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for financial support through projects A06 of SFB 803, A06 of SFB 860, A05 of SFB 937 and through Germany’s Excellence Strategy EXC 2067/1–390729940. We thank the Leibniz Association for financial support through project K76/2017. We also thank B. R. Brueckner for AFM measurements.
Author information
Authors and Affiliations
Contributions
A.G. and N.K. co-designed the project. A.G., A.S. and I.G. performed all the lifetime and defocused imaging measurements. A.G. generated Figs. 2a,c and 3b in the main text. N.K. performed analysis of the defocused imaging data and lifetime data from SLB. A.I.C. carried out the quantum yield measurements with the nanocavity and D.R. performed the corresponding data analysis. S.I. conducted lifetime fitting of single-molecule data and wrote the corresponding section in the Supplementary Information. D.R. helped with writing the MIET routines in Matlab. R.T. designed the DNA origami sample. A.G., N.K. and J.E. carried out the final data analysis, generated all figures (except Figs. 2a,c and 3b) in the main text and wrote the main manuscript. All co-workers were involved with improving the manuscript and writing the Supplementary Information.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
This file contains more information about the work and Supplementary Figs. 1–5.
Rights and permissions
About this article
Cite this article
Ghosh, A., Sharma, A., Chizhik, A.I. et al. Graphene-based metal-induced energy transfer for sub-nanometre optical localization. Nat. Photonics 13, 860–865 (2019). https://doi.org/10.1038/s41566-019-0510-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41566-019-0510-7
This article is cited by
-
Measuring sub-nanometer undulations at microsecond temporal resolution with metal- and graphene-induced energy transfer spectroscopy
Nature Communications (2024)
-
Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy
Light: Science & Applications (2023)
-
An alternative to MINFLUX that enables nanometer resolution in a confocal microscope
Light: Science & Applications (2022)
-
Three-dimensional total-internal reflection fluorescence nanoscopy with nanometric axial resolution by photometric localization of single molecules
Nature Communications (2021)
-
Graphene- and metal-induced energy transfer for single-molecule imaging and live-cell nanoscopy with (sub)-nanometer axial resolution
Nature Protocols (2021)