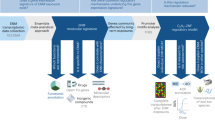
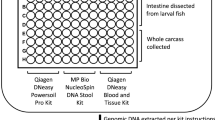
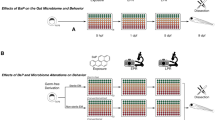
Abstract
Physico-chemical characteristics of engineered nanomaterials are known to be important in determining the impact on organisms but effects are equally dependent upon the characteristics of the organism exposed. Species sensitivity may vary by orders of magnitude, which could be due to differences in the type or magnitude of the biochemical response, exposure or uptake of nanomaterials. Synthesizing conclusions across studies and species is difficult as multiple species are not often included in a study, and differences in batches of nanomaterials, the exposure duration and media across experiments confound comparisons. Here three model species, Danio rerio, Daphnia magna and Chironomus riparius, that differ in sensitivity to lithium cobalt oxide nanosheets are found to differ in immune-response, iron–sulfur protein and central nervous system pathways, among others. Nanomaterial uptake and dissolution does not fully explain cross-species differences. This comparison provides insight into how biomolecular responses across species relate to the varying sensitivity to nanomaterials.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 July 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41565-022-01176-3
References
Klaper, R. D. The known and unknown about the environmental safety of nanomaterials in commerce. Small 16, 2000690 (2020).
Klaper, R., Arndt, D., Bozich, J. & Dominguez, G. Molecular interactions of nanomaterials and organisms: defining biomarkers for toxicity and high-throughput screening using traditional and next-generation sequencing approaches. Analyst 139, 882–895 (2014).
Bondarenko, O. et al. Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review. Arch. Toxicol. 87, 1181–1200 (2013).
Hou, J., Zhou, Y., Wang, C., Li, S. & Wang, X. Toxic effects and molecular mechanism of different types of silver nanoparticles to the aquatic crustacean Daphnia magna. Environ. Sci. Technol. 51, 12868–12878 (2017).
Choi, J. S. & Park, J. W. Molecular characterization and toxicological effects of citrate-coated silver nanoparticles in a terrestrial invertebrate, the earthworm (Eisenia fetida). Mol. Cell. Toxicol. 11, 423–431 (2015).
Scherer, C., Brennholt, N., Reifferscheid, G. & Wagner, M. Feeding type and development drive the ingestion of microplastics by freshwater invertebrates. Sci. Rep. https://doi.org/10.1038/s41598-017-17191-7 (2017).
Rist, S., Baun, A. & Hartmann, N. B. Ingestion of micro- and nanoplastics in Daphnia magna – quantification of body burdens and assessment of feeding rates and reproduction. Environ. Pollut. https://doi.org/10.1016/j.envpol.2017.05.048 (2017).
Spence, R., Gerlach, G., Lawrence, C. & Smith, C. The behaviour and ecology of the zebrafish, Danio rerio. Biol. Rev. Camb. Philos. Soc. 83, 13–34 (2008).
Nath, B. B. Extracellular hemoglobin and environmental stress tolerance in Chironomus larvae. J. Limnol. 77, 104–112 (2018).
Bozich, J., Hang, M., Hamers, R. & Klaper, R. Core chemistry influences the toxicity of multicomponent metal oxide nanomaterials, lithium nickel manganese cobalt oxide, and lithium cobalt oxide to Daphnia magna. Environ. Toxicol. Chem. 36, 2493–2502 (2017).
Niemuth, N. J. et al. Next-generation complex metal oxide nanomaterials negatively impact growth and development in the benthic invertebrate Chironomus riparius upon settling. Environ. Sci. Technol. 53, 3860–3870 (2019).
Burkard, M., Betz, A., Schirmer, K. & Zupanic, A. Common gene expression patterns in environmental model organisms exposed to engineered nanomaterials: a meta-analysis. Environ. Sci. Technol. 54, 335–344 (2020).
Faria, M. et al. Minimum information reporting in bio-nano experimental literature. Nat. Nanotechnol. 13, 777–785 (2018).
Wang, X. et al. Improving cyclic stability of lithium cobalt oxide based lithium ion battery at high voltage by using trimethylboroxine as an electrolyte additive. Electrochim. Acta 173, 804–811 (2015).
Hamers, R. J. Nanomaterials and global sustainability. Acc. Chem. Res. https://doi.org/10.1021/acs.accounts.6b00634 (2017).
McCoole, M. D., Baer, K. N. & Christie, A. E. Histaminergic signaling in the central nervous system of Daphnia and a role for it in the control of phototactic behavior. J. Exp. Biol. 214, 1773–1782 (2011).
Baggelaar, M. P., Maccarrone, M. & van der Stelt, M. 2-Arachidonoylglycerol: a signaling lipid with manifold actions in the brain. Prog. Lipid Res. 71, 1–17 (2018).
Jeong, T.-Y. et al. Effect of β-adrenergic receptor agents on cardiac structure and function and whole-body gene expression in Daphnia magna. Environ. Pollut. https://doi.org/10.1016/j.envpol.2018.06.026 (2018).
Margiotta-Casaluci, L., Owen, S. F., Rand-Weaver, M. & Winter, M. J. Testing the translational power of the zebrafish: an inter-species analysis of responses to cardiovascular drugs. Front. Pharmacol. 10, 893 (2019).
Mendoza, R. P. & Brown, J. M. Engineered nanomaterials and oxidative stress: current understanding and future challenges. Curr. Opin. Toxicol. 13, 74–80 (2019).
Abdelhalim, M. A. K., Qaid, H. A., Al-Mohy, Y. H. & Ghannam, M. M. The protective roles of vitamin E and α-lipoic acid against nephrotoxicity, lipid peroxidation, and inflammatory damage induced by gold nanoparticles. Int. J. Nanomed. 15, 729–734 (2020).
Ha, M. H. & Choi, J. Effects of environmental contaminants on hemoglobin gene expression in Daphnia magna: a potential biomarker for freshwater quality monitoring. Arch. Environ. Contamin. Toxicol. 57, 330–337 (2009).
Prühs, R., Beermann, A. & Schröder, R. The roles of the Wnt-antagonists Axin and Lrp4 during embryogenesis of the red flour beetle Tribolium castaneum. J. Dev. Biol. 5, 10 (2017).
Liang, J. et al. The functions and mechanisms of prefoldin complex and prefoldin-subunits. Cell Biosci. https://doi.org/10.1186/s13578-020-00446-8 (2020).
Serra, A. et al. INSIdE NANO: a systems biology framework to contextualize the mechanism-of-action of engineered nanomaterials. Sci. Rep. 9, 179 (2019).
Newman, M., Ebrahimie, E. & Lardelli, M. Using the zebrafish model for Alzheimer’s disease research. Front. Genet. 5, 189 (2014).
Minegishi, Y., Nakaya, N. & Tomarev, S. I. Mutation in the cebrafish cct2 gene leads to abnormalities of cell cycle and cell death in the retina: a model of CCT2-related Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 59, 995–1004 (2018).
Willardson, B. M. & Howlett, A. C. Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell. Signal. 19, 2417–2427 (2007).
Wu, D., Ma, Y., Cao, Y. & Zhang, T. Mitochondrial toxicity of nanomaterials. Sci. Total Environ. https://doi.org/10.1016/j.scitotenv.2019.134994 (2019).
Nel, A. Toxic potential of materials. Science 311, 622–627 (2007).
Brohi, R. D. et al. Toxicity of nanoparticles on the reproductive system in animal models: a review. Front. Pharmacol. 8, 606 (2017).
Yan, N., Tang, B. Z. & Wang, W.-X. In vivo bioimaging of silver nanoparticle dissolution in the gut environment of zooplankton. ACS Nano https://doi.org/10.1021/acsnano.8b06003 (2018).
Adam, N., Leroux, F., Knapen, D., Bals, S. & Blust, R. The uptake of ZnO and CuO nanoparticles in the water-flea Daphnia magna under acute exposure scenarios. Environ. Pollut. 194, 130–137 (2014).
Lorenz, C. S. et al. Nano-sized Al2O3 reduces acute toxic effects of thiacloprid on the non-biting midge Chironomus riparius. PLoS One 12, e0176356 (2017).
Frouz, J., Lobinske, R. J., Yaqub, A. & Ali, A. Larval gut pH profile in pestiferous Chironomus crassicaudatus and Glyptotendipes paripes (Chironomidae: Diptera) in reference to the toxicity potential of Bacillus thuringiensis serovar israelensis. J. Am. Mosq. Control Assoc. 23, 355–358 (2007).
van Pomeren, M., Brun, N. R., Peijnenburg, W. J. G. M. & Vijver, M. G. Exploring uptake and biodistribution of polystyrene (nano)particles in zebrafish embryos at different developmental stages. Aquat. Toxicol. 190, 40–45 (2017).
Laudadio, E. D., Bennett, J. W., Green, C. M., Mason, S. E. & Hamers, R. J. Impact of phosphate adsorption on complex cobalt oxide nanoparticle dispersibility in aqueous media. Environ. Sci. Technol. 52, 30 (2018).
Niemuth, N. J. et al. Protein Fe–S centers as a molecular target of toxicity of a complex transition metal oxide nanomaterial with downstream impacts on metabolism and growth. Environ. Sci. Technol. 54, 15257–15266 (2020).
Smith, M. & Lazorchak, J. A reformulated, reconstituted water for testing the freshwater amphipod, Hyalella azteca. Environ. Toxicol. Chem. 16, 1229–1233 (1997).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. https://doi.org/10.1038/nbt.1883 (2011).
The UniProt Consortium, UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics https://doi.org/10.1186/1471-2105-10-421 (2009).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Schmidt, H. et al. A high-quality genome assembly from short and long reads for the non-biting midge Chironomus riparius (Diptera). G3 (Bethesda) https://doi.org/10.1534/g3.119.400710 (2020).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K.-A. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. https://doi.org/10.1371/journal.pcbi.1005752 (2017).
Chiesa, M., Colombo, G. I. & Piacentini, L. DaMiRseq—an R/Bioconductor package for data mining of RNA-Seq data: normalization, feature selection and classification. Bioinformatics 34, 1416–1418 (2018).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Mi, H. et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, 191–198 (2019).
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant No. CHE-2001611, the NSF Center for Sustainable Nanotechnology (R.J.H and R.D.K.). The Center for Sustainable Nanotechnology is part of the Centers for Chemical Innovation Program. This project used the UWM Great Lakes Genomics Center sequencing and bioinformatics services. UWM Institutional Animal Care and Use Committee protocols followed 20-21 no. 01 and 20-21 no. 50.
Author information
Authors and Affiliations
Contributions
B.J.C., N.J.N. and R.D.K. conceived the experiment and its design. B.J.C., N.J.N. and E.B. carried out the LCO nanosheet exposures. A.S. prepared the RNA-Seq libraries. O.M. and A.A.M. conducted bioinformatic quality-control analysis and RNA-Seq data analysis. B.J.C. carried out additional downstream analyses, including PLS-DA, pathway classification and enrichment analysis. E.D.L. and R.J.H. provided nanomaterial synthesis and characterization. Y.S. and J.C.W. carried out elemental analysis. B.J.C. and R.D.K. wrote and edited the paper, with contributions and support from all co-authors. Research was supervised by R.D.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary graphical abstract, Figs. 1–5, Tables 1 and 2, methods and discussion.
Rights and permissions
About this article
Cite this article
Curtis, B.J., Niemuth, N.J., Bennett, E. et al. Cross-species transcriptomic signatures identify mechanisms related to species sensitivity and common responses to nanomaterials. Nat. Nanotechnol. 17, 661–669 (2022). https://doi.org/10.1038/s41565-022-01096-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-022-01096-2
This article is cited by
-
A singular plasmonic-thermoelectric hollow nanostructure inducing apoptosis and cuproptosis for catalytic cancer therapy
Nature Communications (2024)
-
An ancestral molecular response to nanomaterial particulates
Nature Nanotechnology (2023)