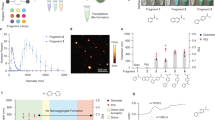
Abstract
Nanoformulations of therapeutic drugs are transforming our ability to effectively deliver and treat a myriad of conditions. Often, however, they are complex to produce and exhibit low drug loading, except for nanoparticles formed via co-assembly of drugs and small molecular dyes, which display drug-loading capacities of up to 95%. There is currently no understanding of which of the millions of small-molecule combinations can result in the formation of these nanoparticles. Here we report the integration of machine learning with high-throughput experimentation to enable the rapid and large-scale identification of such nanoformulations. We identified 100 self-assembling drug nanoparticles from 2.1 million pairings, each including one of 788 candidate drugs and one of 2,686 approved excipients. We further characterized two nanoparticles, sorafenib–glycyrrhizin and terbinafine–taurocholic acid both ex vivo and in vivo. We anticipate that our platform can accelerate the development of safer and more efficacious nanoformulations with high drug-loading capacities for a wide range of therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used in this study were extracted from publicly available sources from the DrugBank website (version 5.0, drugbank.ca), the FDA (GRAS version June 2016 and IIG version 0716 UNII 362O9ITL9D datasets, www.fda.gov) and the Shoichet laboratory (bkslab.org/takeaways/aggregator_hts). The processed and curated data are made available through the GitHub repository at https://github.com/DanReker/CoAggregators.
Code availability
All code for this study to perform simulations and machine learning is available through the GitHub repository at https://github.com/DanReker/CoAggregators.
References
Hopkins, A. L., Keserü, G. M., Leeson, P. D., Rees, D. C. & Reynolds, C. H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 13, 105–121 (2014).
Irwin, J. J. et al. An aggregation advisor for ligand discovery. J. Med. Chem. 58, 7076–7087 (2015).
Reker, D., Bernardes, G. J. L. & Rodrigues, T. Computational advances in combating colloidal aggregation in drug discovery. Nat. Chem. 11, 402–418 (2019).
Owen, S. C., Doak, A. K., Wassam, P., Shoichet, M. S. & Shoichet, B. K. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol. 7, 1429–1435 (2012).
Kipp, J. E. The role of solid nanoparticle technology in the parenteral delivery of poorly water-soluble drugs. Int. J. Pharm. 284, 109–122 (2004).
Mcdonald, T. O. et al. Antiretroviral solid drug nanoparticles with enhanced oral bioavailability: production, characterization, and in vitro-in vivo correlation. Adv. Healthc. Mater. 3, 400–411 (2014).
Govender, T., Stolnik, S., Garnett, M. C., Illum, L. & Davis, S. S. PLGA nanoparticles prepared by nanoprecipitation: drug loading and release studies of a water soluble drug. J. Control. Release 57, 171–185 (1999).
Westesen, K., Bunjes, H. & Koch, M. H. Physicochemical characterization of lipid nanoparticles and evaluation of their drug loading capacity and sustained release potential. J. Control. Release 48, 223–236 (1997).
Reker, D. et al. ‘Inactive’ ingredients in oral medications. Sci. Transl. Med. 11, eaau6753 (2019).
McLaughlin, C. K. et al. Stable colloidal drug aggregates catch and release active enzymes. ACS Chem. Biol. 11, 992–1000 (2016).
Shamay, Y. et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater. 17, 361–368 (2018).
Inactive Ingredient Search for Approved Drug Products (FDA, 2016); https://www.accessdata.fda.gov/scripts/cder/iig/
Feng, B. Y., Shelat, A., Dorman, T. N., Guy, R. K. & Shoichet, B. K. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148 (2005).
SCOGS (Select Committee on GRAS Substances) (FDA, 2016); https://www.accessdata.fda.gov/scripts/fdcc/?set=SCOGS
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Reker, D. et al. Machine learning uncovers food- and excipient-drug interactions. Cell Rep. 30, 3710–3716.e4 (2020).
Reker, D., Schneider, P. & Schneider, G. Multi-objective active machine learning rapidly improves structure-activity models and reveals new protein-protein interaction inhibitors. Chem. Sci. 7, 3919–3927 (2016).
Gregori-Puigjane, E. & Mestres, J. A ligand-based approach to mining the chemogenomic space of drugs. Comb. Chem. High T. Scr. 11, 669–676 (2008).
Wildman, S. A. & Crippen, G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comp. Sci. 39, 868–873 (1999).
Chuang, K. V. & Keiser, M. J. Adversarial controls for scientific machine learning. ACS Chem. Biol. 13, 2819–2821 (2018).
Test no. 318: dispersion stability of nanomaterials in simulated environmental media. OECD Guidelines for the Testing of Chemicals, Section 3 https://doi.org/10.1787/9789264284142-en (2017).
Lipner, S. R. & Scher, R. K. Onychomycosis: treatment and prevention of recurrence. J. Am. Acad. Dermatol. 80, 853–867 (2019).
McClellan, K. J., Wiseman, L. R. & Markham, A. Terbinafine. Drugs 58, 179–202 (1999).
Matteucci, M. E., Hotze, M. A., Johnston, K. P. & Williams, R. O. Drug nanoparticles by antisolvent precipitation: mixing energy versus surfactant stabilization. Langmuir 22, 8951–8959 (2006).
Ganesh, A. N. et al. Colloidal drug aggregate stability in high serum conditions and pharmacokinetic consequence. ACS Chem. Biol. 14, 751–757 (2019).
Jayatilake, J. A. M. S., Tilakaratne, W. M. & Panagoda, G. J. Candidal onychomycosis: a mini-review. Mycopathologia 168, 165–173 (2009).
Cabral, H. et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 6, 815–823 (2011).
Wong, C. et al. Multistage nanoparticle delivery system for deep penetration into tumor tissue. Proc. Natl Acad. Sci. USA 108, 2426–2431 (2011).
Pavlović, N. et al. Bile acids and their derivatives as potential modifiers of drug release and pharmacokinetic profiles. Front. Pharmacol. 9, 1283 (2018).
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
El-Serag, H. B. & Mason, A. C. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 340, 745–750 (1999).
Isbrucker, R. A. A. & Burdock, G. A. A. Risk and safety assessment on the consumption of Licorice root (Glycyrrhiza sp.), its extract and powder as a food ingredient, with emphasis on the pharmacology and toxicology of glycyrrhizin. Regul. Toxicol. Pharm. 46, 167–192 (2006).
Basso, U., Brunello, A., Bertuzzi, A. & Santoro, A. Sorafenib is active on lung metastases from synovial sarcoma. Ann. Oncol. 20, 386–387 (2009).
Chaparro, M., González Moreno, L., Trapero-Marugán, M., Medina, J. & Moreno-Otero, R. Review article: pharmacological therapy for hepatocellular carcinoma with sorafenib and other oral agents. Aliment. Pharm. Ther. 28, 1269–1277 (2008).
Zhong, J. et al. Meloxicam combined with sorafenib synergistically inhibits tumor growth of human hepatocellular carcinoma cells via ER stress-related apoptosis. Oncol. Rep. 34, 2142–2150 (2015).
Auffan, M. et al. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. https://doi.org/10.1038/nnano.2009.242 (2009).
Lin, A. et al. Glycyrrhizin surface-modified chitosan nanoparticles for hepatocyte-targeted delivery. Int. J. Pharm. 359, 247–253 (2008).
Tward, A. D. et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc. Natl Acad. Sci. USA 104, 14771–14776 (2007).
Bogorad, R. L. et al. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat. Commun. 5, 3869 (2014).
Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006).
Sharpless, N. E. & DePinho, R. A. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 5, 741–754 (2006).
Waidely, E., Al-Yuobi, A. R. O., Bashammakh, A. S., El-Shahawi, M. S. & Leblanc, R. M. Serum protein biomarkers relevant to hepatocellular carcinoma and their detection. Analyst 141, 36–44 (2016).
Lee, C. H. et al. Protective mechanism of glycyrrhizin on acute liver injury induced by carbon tetrachloride in mice. Biol. Pharm. Bull. 30, 1898–1904 (2007).
Gelderblom, H., Verweij, J., Nooter, K., Sparreboom, A. & Cremophor, E. L. The drawbacks and advantages of vehicle selection for drug formulation. Eur. J. Cancer 37, 1590–1598 (2001).
Singh, A., Iyer, A. K. & Amiji, M. M. in Handbook of Nanobiomedical Research (ed. Torchilin, V.) 199–233 (World Scientific Publishing Company, 2014); https://doi.org/10.1142/9789814520652_0006
Pohjala, L., Tammela, P., Pohjala, L. & Tammela, P. Aggregating behavior of phenolic compounds—a source of false bioassay results? Molecules 17, 10774–10790 (2012).
Traverso, G. & Langer, R. Perspective: special delivery for the gut. Nature 519, S19 (2015).
Cheng, C. J. & Saltzman, W. M. Nanomedicine: downsizing tumour therapeutics. Nat. Nanotechnol. 7, 346–347 (2012).
Acknowledgements
D.R. is a Swiss National Science Foundation Fellow (grants P2EZP3_168827 and P300P2_177833). Y.R. is grateful to the MIT Skoltech Initiative for financial support. A.R.K. is grateful to the PhRMA foundation postdoctoral fellowship for financial support. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the High Throughput Sciences Facility, Peterson (1957) Nanotechnology Materials Core Facility, the Animal Imaging and Preclinical Testing core, and the Hope Babette Tang (1983) Histology Facility. This work was supported in part by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute and by the NIH grant EB000244. We are grateful to OpenEye for providing us with an OpenEye Academic License. We are grateful to H. Mazdiyasni for technical support and to M. Jimenez for providing access to C. albicans and helpful discussions throughout the study.
Author information
Authors and Affiliations
Contributions
D.R., R.L. and G.T. conceived the study. D.R., Y.R., A.R.K., R.L. and G.T. designed experiments. D.R. and J.W.Y. performed in silico experiments. D.R., R.C., J.W.Y., N.N., R.M.Z., T.E. and J.L. performed in vitro experiments. D.R., R.C. and A.G. performed in vivo experiments. Y.R., A.R.K., T.v.E., A.L.-J., C.K.S. and J.H.C. supported in vitro experiments. Y.R., A.R.K., E.M.S., D.L., J.C., S.M.T., K.I., P.C. and A.M.H. supported in vivo experiments. D.Y. performed TEM imaging, and K.H., A.L. and J.R. performed pharmaceutical analytics. D.R., R.L. and G.T. wrote the manuscript with contributions from the other authors. All authors approved the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
D.R., Y.R., A.R.K., R.C., J.W.Y., N.N., A.G., R.M.Z., R.L. and G.T. are co-inventors on multiple patent applications describing novel nanoformulation systems and interactions between excipients and drugs. D.R. acts as a mentor for the German Accelerator Life Sciences and acts as a scientific consultant for pharmaceutical and biotechnology companies.
Additional information
Peer review information Nature Nanotechnology thanks Michael Keiser, Jie Zheng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Methods, Tables 1–13 and Notes 1–7.
Rights and permissions
About this article
Cite this article
Reker, D., Rybakova, Y., Kirtane, A.R. et al. Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat. Nanotechnol. 16, 725–733 (2021). https://doi.org/10.1038/s41565-021-00870-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-00870-y
This article is cited by
-
Strategies for non-viral vectors targeting organs beyond the liver
Nature Nanotechnology (2024)
-
The landscape of small-molecule prodrugs
Nature Reviews Drug Discovery (2024)
-
Application of ensemble machine learning approach to assess the factors affecting size and polydispersity index of liposomal nanoparticles
Scientific Reports (2023)
-
Fragment-based drug nanoaggregation reveals drivers of self-assembly
Nature Communications (2023)
-
Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences
Nature Biomedical Engineering (2023)