Abstract
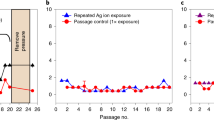
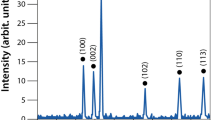
Silver nanoparticles have already been successfully applied in various biomedical and antimicrobial technologies and products used in everyday life. Although bacterial resistance to antibiotics has been extensively discussed in the literature, the possible development of resistance to silver nanoparticles has not been fully explored. We report that the Gram-negative bacteria Escherichia coli 013, Pseudomonas aeruginosa CCM 3955 and E. coli CCM 3954 can develop resistance to silver nanoparticles after repeated exposure. The resistance stems from the production of the adhesive flagellum protein flagellin, which triggers the aggregation of the nanoparticles. This resistance evolves without any genetic changes; only phenotypic change is needed to reduce the nanoparticles’ colloidal stability and thus eliminate their antibacterial activity. The resistance mechanism cannot be overcome by additional stabilization of silver nanoparticles using surfactants or polymers. It is, however, strongly suppressed by inhibiting flagellin production with pomegranate rind extract.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wong, K. K. Y. & Liu, X. L. Silver nanoparticles—the real ‘silver bullet’ in clinical medicine? MedChemComm 1, 125–131 (2010).
Livermore, D. M. Fourteen years in resistance. Int. J. Antimicrob. Agents 39, 283–294 (2012).
May, M. Antibiotics. Nature 509, S1 (2014).
Hede, K. An infectious arms race. Nature 509, S2–S3 (2014).
Cui, L. et al. In situ study of the antibacterial activity and mechanism of action of silver nanoparticles by surface-enhanced Raman spectroscopy. Anal. Chem. 85, 5436–5443 (2013).
Lara, H. H., Ayala-Núñez, N. V., Ixtepan Turrent, L. del C. & Rodríguez Padilla, C. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiol. Biotechnol. 26, 615–621 (2010).
Li, W.-R. et al. Antibacterial effect of silver nanoparticles on Staphylococcus aureus. BioMetals 24, 135–141 (2011).
Morones, J. R. et al. The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346–2353 (2005).
Panáček, A. et al. Silver colloid nanoparticles: synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 110, 16248–16253 (2006).
Panáček, A. et al. Strong and nonspecific synergistic antibacterial efficiency of antibiotics combined with silver nanoparticles at very low concentrations showing no cytotoxic effect. Molecules 21, 26 (2016).
Panáček, A. et al. Silver nanoparticles strongly enhance and restore bactericidal activity of inactive antibiotics against multiresistant Enterobacteriaceae. Colloids Surf. B Biointerfaces 142, 392–399 (2016).
Brown, A. N. et al. Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl. Environ. Microbiol. 78, 2768–2774 (2012).
Sharma, V. K., Yngard, R. A. & Lin, Y. Silver nanoparticles: green synthesis and their antimicrobial activities. Adv. Colloid Interface Sci. 145, 83–96 (2009).
Morones-Ramirez, J. R., Winkler, J. A., Spina, C. S. & Collins, J. J. Silver enhances antibiotic activity against Gram-negative bacteria. Sci. Transl. Med. 5, 190ra81 (2013).
Lemire, J. A., Harrison, J. J. & Turner, R. J. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384 (2013).
Haefeli, C., Franklin, C. & Hardy, K. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158, 389–392 (1984).
Li, X. Z., Nikaido, H. & Williams, K. E. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J. Bacteriol. 179, 6127–6132 (1997).
Gupta, A., Matsui, K., Lo, J.-F. & Silver, S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5, 183–188 (1999).
Nies, D. H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339 (2003).
Silver, S. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27, 341–353 (2003).
Silver, S., Phung, L. T. & Silver, G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 33, 627–634 (2006).
Graves, J. L. Jr et al. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 6, 42 (2015).
Losasso, C. et al. Antibacterial activity of silver nanoparticles: sensitivity of different Salmonella serovars. Front. Microbiol. 5, 227 (2014).
Gunawan, C., Teoh, W. Y., Marquis, C. P. & Amal, R. Induced adaptation of Bacillus sp. to antimicrobial nanosilver. Small 9, 3554–3560 (2013).
Kvítek, L. et al. Effect of surfactants and polymers on stability and antibacterial activity of silver nanoparticles (NPs). J. Phys. Chem. C 112, 5825–5834 (2008).
Sivera, M. et al. Silver nanoparticles modified by gelatin with extraordinary pH stability and long-term antibacterial activity. PLoS One 9, e103675 (2014).
Hunter, R. J. in Foundations of Colloid Science 2nd edn, 601–603 (Oxford Univ. Press, New York, 2001).
Bardy, S. L., Ng, S. Y. M. & Jarrell, K. F. Prokaryotic motility structures. Microbiology 149, 295–304 (2003).
Metlina, A. L. Bacterial and archaeal flagella as prokaryotic motility organelles. Biochemistry (Moscow) 69, 1203–1212 (2004).
Hayashi, F. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001).
Friedlander, R. S. et al. Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc. Natl Acad. Sci. USA 110, 5624–5629 (2013).
Haiko, J. & Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267 (2013).
Pratt, L. A. & Kolter, R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293 (1998).
Asadishad, B., Hidalgo, G. & Tufenkji, N. Pomegranate materials inhibit flagellin gene expression and flagellar-propelled motility of uropathogenic Escherichia coli strain CFT073. FEMS Microbiol. Lett. 334, 87–94 (2012).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. O. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Petrovská, B. et al. Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet. Genome Res. 143, 78–86 (2014).
Koboldt, D. C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Acknowledgements
We acknowledge the support provided by projects LO1305 and LM2015073 of the Ministry of Education, Youth and Sports of the Czech Republic, the Czech Science Foundation (project no.15–22248S), the Ministry of Health of the Czech Republic (AZV VES 15-27726A) and the Internal Grants of Palacký University in Olomouc (IGA_PrF_2015_022 and IGA_LF_2016_022).
Author information
Authors and Affiliations
Contributions
A.P. and L.K. conceived the project and designed the experiments dealing with synthesis of silver NPs. M.S. and R.P. carried out the experiments dealing with synthesis and stabilizations of silver NPs including their characterizations. R.V. and M.K. designed and carried out the microbiological experiments. M.R. carried out the experiments dealing with genomic analysis. F.D. and M.S. carried out the experiments dealing with proteomic analysis and viability tests. O.T. was responsible for HRTEM characterizations. R.Z. contributed clarifications and guidance on the manuscript. A.P. and R.Z. wrote the paper with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures 1–8 and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Panáček, A., Kvítek, L., Smékalová, M. et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nature Nanotech 13, 65–71 (2018). https://doi.org/10.1038/s41565-017-0013-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-017-0013-y
This article is cited by
-
Antimicrobial Metal and Metal Oxide Nanoparticles in Bone Tissue Repair
Biomedical Materials & Devices (2024)
-
Green One-Step Synthesis of Silver Nanoparticles Obtained from Schinus areira Leaf Extract: Characterization and Antibacterial Mechanism Analysis
Applied Biochemistry and Biotechnology (2024)
-
Analytical methods for assessing antimicrobial activity of nanomaterials in complex media: advances, challenges, and perspectives
Journal of Nanobiotechnology (2023)
-
The efficacy of biosynthesized silver nanoparticles against Pseudomonas aeruginosa isolates from cystic fibrosis patients
Scientific Reports (2023)
-
Studying the Photocatalytic Characteristics of Pd Nanoparticles Created by Second Harmonic Laser Ablation
Plasmonics (2023)