Abstract
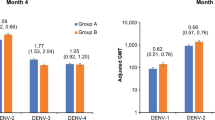
Dengue human infection models present an opportunity to explore the potential of a vaccine, anti-viral or immuno-compound for clinical benefit in a controlled setting. Here we report the outcome of a phase 1 open-label assessment of a low-dose dengue virus 3 (DENV-3) challenge model (NCT04298138), in which nine participants received a subcutaneous inoculation with 0.5 ml of a 1.4 × 103 plaque-forming unit per ml suspension of the attenuated DENV-3 strain CH53489. The primary and secondary endpoints of the study were to assess the safety of this DENV-3 strain in healthy flavivirus-seronegative individuals. All participants developed RNAaemia within 7 days after inoculation with peak titre ranging from 3.13 × 104 to 7.02 × 108 genome equivalents per ml. Solicited symptoms such as fever and rash, clinical laboratory abnormalities such as lymphopenia and thrombocytopenia, and self-reported symptoms such as myalgia were consistent with mild-to-moderate dengue in all volunteers. DENV-3-specific seroconversion and memory T cell responses were observed within 14 days after inoculation as assessed by enzyme-linked immunosorbent assay and interferon-gamma-based enzyme-linked immunospot. RNA sequencing and serum cytokine analysis revealed anti-viral responses that overlapped with the period of viraemia. The magnitude and frequency of clinical and immunologic endpoints correlated with an individual’s peak viral titre.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study, including participant-level source data, are available within this article, in its Supplementary Information files or in a publicly accessible database. RNAseq gene expression data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO GSE216328). The DENV strain used for this study was made available by the US Army Medical Research and Development Command under a material transfer agreement with the SUNY–UMU and SUNY Research Foundation. Requests for the challenge strain should be addressed to the US Army Medical Research and Development Command (heather.l.friberg-robertson.civ@health.mil). Please anticipate a response within 2 weeks.
References
Bhatt, S. et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013).
Shepard, D. S., Coudeville, L., Halasa, Y. A., Zambrano, B. & Dayan, G. H. Economic impact of dengue illness in the Americas. Am. J. Trop. Med. Hyg. 84, 200–207 (2011).
Guzman, M. G., Gubler, D. J., Izquierdo, A., Martinez, E. & Halstead, S. B. Dengue infection. Nat. Rev. Dis. Primers 2, 16055 (2016).
Gubler, D. J. Aedes aegypti and Aedes aegypti-borne disease control in the 1990s: top down or bottom up. Charles Franklin Craig lecture. Am. J. Trop. Med. Hyg. 40, 571–578 (1989).
Iwamura, T., Guzman-Holst, A. & Murray, K. A. Accelerating invasion potential of disease vector Aedes aegypti under climate change. Nat. Commun. 11, 2130 (2020).
Facts sheet Dengue. World Mosquito Program https://www.worldmosquitoprogram.org/sites/default/files/2020-11/WMP%20dengue_0.pdf (2022).
Anderson, K. B. et al. A shorter time interval between first and second dengue infections is associated with protection from clinical illness in a school-based cohort in Thailand. J. Infect. Dis. 209, 360–368 (2014).
Montoya, M. et al. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl. Trop. Dis. 7, e2357 (2013).
Guzman, M. G., Alvarez, M. & Halstead, S. B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 158, 1445–1459 (2013).
Rajapakse, S., de Silva, N. L., Weeratunga, P., Rodrigo, C. & Fernando, S. D. Prophylactic and therapeutic interventions for bleeding in dengue: a systematic review. Trans. R. Soc. Trop. Med. Hyg. 111, 433–439 (2017).
Wong, J. G., Thein, T. L., Leo, Y. S., Pang, J. & Lye, D. C. Identifying adult dengue patients at low risk for clinically significant bleeding. PLoS ONE 11, e0148579 (2016).
Capeding, M. R. et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384, 1358–1365 (2014).
Villar, L. et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. N. Engl. J. Med. 372, 113–123 (2015).
World Health, O. Dengue vaccine: WHO position paper, July 2016—recommendations. Vaccine 35, 1200–1201 (2017).
Biswal, S. et al. Efficacy of a tetravalent dengue vaccine in healthy children and adolescents. N. Engl. J. Med. 381, 2009–2019 (2019).
Biswal, S. et al. Efficacy of a tetravalent dengue vaccine in healthy children aged 4-16 years: a randomised, placebo-controlled, phase 3 trial. Lancet 395, 1423–1433 (2020).
Takeda’s QDENGA (dengue tetravalent vaccine (live, attenuated)) approved in Indonesia for use regardless of prior dengue exposure. Takeda https://www.takeda.com/newsroom/newsreleases/2022/takedas-qdenga-dengue-tetravalentvaccine-live-attenuated-approved-in-indonesia-for-useregardless-of-prior-dengue-exposure/ (2022).
Thomas, S. J. & Endy, T. P. Current issues in dengue vaccination. Curr. Opin. Infect. Dis. 26, 429–434 (2013).
Low, J. G., Ooi, E. E. & Vasudevan, S. G. Current status of dengue therapeutics research and development. J. Infect. Dis. 215, S96–S102 (2017).
Thomas, S. J. Is new dengue vaccine efficacy data a relief or cause for concern? NPJ Vaccines 8, 55 (2023).
Endy, T. P. et al. A phase 1, open-label assessment of a dengue virus-1 live virus human challenge strain. J. Infect. Dis. 223, 258–267 (2021).
Mammen, M. P. et al. Evaluation of dengue virus strains for human challenge studies. Vaccine 32, 1488–1494 (2014).
Sun, W. et al. Experimental dengue virus challenge of human subjects previously vaccinated with live attenuated tetravalent dengue vaccines. J. Infect. Dis. 207, 700–708 (2013).
Waickman, A. T. et al. Evolution of inflammation and immunity in a dengue virus 1 human infection model. Sci. Transl. Med. 14, eabo5019 (2022).
Waickman, A. T. et al. Transcriptional and clonal characterization of B cell plasmablast diversity following primary and secondary natural DENV infection. EBioMedicine 54, 102733 (2020).
Kirkpatrick, B. D. et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci. Transl. Med. 8, 330ra336 (2016).
Hou, R. et al. The innate immune response following multivalent dengue vaccination and implications for protection against dengue challenge. JCI Insight https://doi.org/10.1172/jci.insight.157811 (2022).
Nivarthi, U. K. et al. Longitudinal analysis of acute and convalescent B cell responses in a human primary dengue serotype 2 infection model. EBioMedicine 41, 465–478 (2019).
Hanley, J. P. et al. Immunotranscriptomic profiling the acute and clearance phases of a human challenge dengue virus serotype 2 infection model. Nat. Commun. 12, 3054 (2021).
Sabin, A. B. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1, 30–50 (1952).
McCracken, M. K. et al. Route of inoculation and mosquito vector exposure modulate dengue virus replication kinetics and immune responses in rhesus macaques. PLoS Negl.Trop. Dis. 14, e0008191 (2020).
Dengue 3 human infection model (DENV-3). ClinicalTrials.gov https://www.clinicaltrials.gov/study/NCT04298138 (2022).
Houng, H. S., Chung-Ming Chen, R., Vaughn, D. W. & Kanesa-thasan, N. Development of a fluorogenic RT–PCR system for quantitative identification of dengue virus serotypes 1–4 using conserved and serotype-specific 3′ noncoding sequences. J. Virol. Methods 95, 19–32 (2001).
Kraus, A. A., Messer, W., Haymore, L. B. & de Silva, A. M. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J. Clin. Microbiol. 45, 3777–3780 (2007).
de Alwis, R. et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl Acad. Sci. USA 109, 7439–7444 (2012).
Berry, A. S. F. et al. An open-source toolkit to expand bioinformatics training in infectious diseases. mBio 12, e0121421 (2021).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Erratum: near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 888 (2016).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
Bolotin, D. A. et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 35, 908–911 (2017).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Lee, D. W. et al. BRILIA: integrated tool for high-throughput annotation and lineage tree assembly of B-cell repertoires. Front. Immunol. 7, 681 (2016).
Acknowledgements
We gratefully acknowledge excellent technical assistance provided by K. Gentile of the UMU Molecular Analysis Core, L. Phelps of the SUNY–UMU Flow Cytometry Core, H. Chanatry and the members of the Institute for Global Health and Translational Science of SUNY–UMU. We also acknowledge C. Rooney from the US Army. We also wish to thank all the study participants for making this study possible. The following reagents were obtained through BEI Resources, NIAID, NIH: Peptide Array, DENV-3 Sleman/1978, E protein (NR-511), DENV-3 Philippines/H87/1956, E protein (NR-9228), DENV-3 Philippines/H87/1956, NS1 protein (NR-2753), DENV-3 Philippines/H87/1956, NS3 protein (NR-2754), DENV-3 Philippines/H87/1956 and NS5 protein (NR-4204). The opinions or assertions contained herein are the private views of the authors and are not to be construed as reflecting the official views of the US Army or the US Department of Defense. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The investigators have adhered to the policies for protection of human participants as prescribed in AR 70-25. The sponsor was involved in the design and conduct of the study and in the collection, management, analysis and interpretation of the data. All authors had full access to the data in the study. Funding: funding for this research was provided by the Department of Defense, Medical Research and Material Command, the Military Infectious Disease Research Program (S.J.T. and T.P.E.) and the State of New York (A.T.W.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: L.A.W., M.D.K., R.G.J., T.P.E. and S.J.T. Formal analysis: L.A.W. and A.T.W. Funding acquisition: H.F, R.G.J., T.P.E. and S.J.T. Investigation: A.T.W., J.Q.L., K.N., H.S.F., M.W., C.G., M.D.K., L.A.W., T.P.E. and S.J.T. Resources: H.F., R.G.J. and J.R.C. Visualization: K.N. and A.T.W. Writing—original draft: A.T.W. and S.J.T. Writing—review and editing: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Anna Durbin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 NS1 opsonization assay.
a) Sequence and annotation of the DENV-3 NS1 expression construct used in this study. Yellow = signal peptide, green = NS1, blue = NS2A. b) Anti-NS1 staining mAb staining (clone FE8) of parental CEM.NKR cell line and DENV-3 NS1 expressing CEM.NKR cells. Unstained cells shown in blue, FE8 stained shown in red c) Representative NS1 opsonizing activity of serum collected at days 0, 24, and 90 days post DENV-3 challenge. Unstained cells shown in blue, serum stained shown in red.
Extended Data Fig. 2 DENV serology day 90 post challenge.
a) Quantification of DENV-1, −2, −3 and −4 E binding IgG antibody levels on day 90 post challenge using a multiplex antigen array. b) Quantification of DENV-1, −2, −3 and −4 E neutralizing antibody titers on 90 post challenge by FlowNT. * p < 0.05 paired one-way ANOVA with correction for multiple comparisons. Data are presented as mean values +/- SEM. n = 9 biologically independent samples.
Extended Data Fig. 3
Representative IFN-γ ELISPOT images.
Extended Data Fig. 4 Gene module identification from RNA-seq data.
Identification of gene modules with coordinated expression changes following DENV-3 challenge. a) Gene modules identified performing differential gene expression analysis between A) days 0 and 6 post infection, b) days 0 and 8 post infection, c) days 0 and 10 post infection, and d) days 0 and 14 post infection.
Extended Data Fig. 5 Select extended correlation analysis.
Correlation analysis between peak virama titers and T cell responses day 90 post infection. Relationship between peak viremia and a) DENV-3 E protein ELISPOT readout, b) DENV-3 NS1 protein ELISPOT readout, c) DENV-3 NS3 protein ELISPOT readout, d) DENV-3 NS5 protein ELISPOT readout on day 90 post.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7 and Tables 1–6.
Supplementary Data 1
RNAseq differential gene expression data.
Supplementary Data 2
Study protocol.
Source data
Source Data
Statistical source data for Figs. 1–6 and Extended Figs. 1–2 and 5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Waickman, A.T., Newell, K., Lu, J.Q. et al. Low-dose dengue virus 3 human challenge model: a phase 1 open-label study. Nat Microbiol (2024). https://doi.org/10.1038/s41564-024-01668-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41564-024-01668-z