Abstract
DNA-amplicon-based microbiota profiling can estimate species diversity and abundance but cannot resolve genetic differences within individuals of the same species. Here we report the development of modular bacterial tags (MoBacTags) encoding DNA barcodes that enable tracking of near-isogenic bacterial commensals in an array of complex microbiome communities. Chromosomally integrated DNA barcodes are then co-amplified with endogenous marker genes of the community by integrating corresponding primer binding sites into the barcode. We use this approach to assess the contributions of individual bacterial genes to Arabidopsis thaliana root microbiota establishment with synthetic communities that include MoBacTag-labelled strains of Pseudomonas capeferrum. Results show reduced root colonization for certain mutant strains with defects in gluconic-acid-mediated host immunosuppression, which would not be detected with traditional amplicon sequencing. Our work illustrates how MoBacTags can be applied to assess scaling of individual bacterial genetic determinants in the plant microbiota.
Similar content being viewed by others
Main
Plants are inhabited by taxonomically structured multi-kingdom microbial communities referred to as plant microbiota. Members of the microbiota provide beneficial services to the plant host1, including mobilization of nutrients2,3, indirect pathogen protection4,5,6 and abiotic stress tolerance7,8. Quantitative and cultivation-independent analysis of these microbial communities using marker genes, such as the 16S rRNA gene of bacteria or internal transcribed spacer (ITS) regions of fungi, typically relies on the detection of natural nucleotide polymorphisms in hypervariable regions of the markers, thereby defining distinct microbial taxa. Computational analysis of marker-gene-based amplicon DNA sequencing data generated using PCR with marker-gene-specific primers has shifted from clustering similar reads into operational taxonomic units to error-correction approaches that account for individual amplicon sequence variants9,10,11. Despite an increased taxonomic resolution of bacterial communities through classification by amplicon sequence variants, profiling based on 16S rRNA is still unable to capture the true within-phylotype genetic variation of microbial communities. Strains with identical marker gene sequences may comprise a bacterial population with other polymorphic loci associated with beneficial or detrimental plant traits, as shown for Rhizobiales and Pseudomonas lineages12,13,14. In such cases, only cumulative relative 16S rRNA abundances originating from multiple strains represented by a phylotype can be retrieved (Fig. 1a). This imposes limitations on functional microbiota studies, as the bacterial traits often vary in a strain-specific manner. Studies on genetic variation within a bacterial phylotype are therefore typically limited to host mono-associations with cultured bacterial or fungal strains, and competition experiments with antibiotic markers15, or depend on DNA sequencing of strain-differentiating amplicons restricted to a particular taxon16. Furthermore, amplification of endogenous marker sequences does not allow differentiation between wild-type and mutant strains in a community context, limiting the application of microbial genetics in gnotobiotic systems.
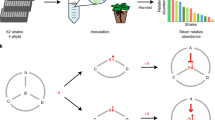
a, Principles of MoBacTag DNA barcodes for discriminating bacterial strains encoding identical 16S rRNA sequences during amplicon sequencing for microbiota profiling and for quantifying microbial load using MoBacTag-based spike-in plasmids. b, Schematic representation of the MoBacTag. Each vector encodes 1 of the 20 possible combinations of the 4 antibiotics with the 5 fluorescent markers. The MoBacTag fragment (grey bar) is chromosomally integrated downstream of the glucosamine-6-phosphate synthetase (glmS) gene. The synthetic DNA tag is flanked by primer binding sites for amplification of bacterial 16S, fungal ITS, plant ITS or barcode sequences. Plasmids encoding only the synthetic DNA tag were further used as spike plasmids. Tn7R and Tn7L, right and left Tn7 borders, respectively; SpSmR, spectinomycin and streptomycin resistance; KnR, kanamycin resistance; TcR, tetracycline resistance; GmR, gentamycin resistance; Protac, synthetic bacterial hybrid promoter; sacB, levansucrase gene conferring sucrose sensitivity; tagBFP, blue fluorescent protein; AmCyan1, cyan fluorescent protein; far-red FP, far red fluorescent protein. The elements are not drawn to scale. c, Oligonucleotide composition of the synthetic DNA tags. The upper panel shows the number of oligonucleotides integrated into the DNA barcode tags. Grey colours represent two different assembly batches. The lower panel shows heat-map-visualized frequencies of individual oligonucleotides (a–j) in forward or reverse orientation at each position (p1, p2, p3, p4 and p5) of the array of ligated oligonucleotides within the DNA barcode tag. d, Integration strategy of preassembled DNA barcode tags into MoBacTag recipient vectors. Preassembled DNA barcode tags are inserted between primer binding sites in exchange for the dominant negative selection marker ccdB. cmR, chloramphenicol resistance.
Cellular barcodes can overcome resolution limitations by labelling strains or individual cells with unique DNA sequences, here referred to as DNA barcodes (Fig. 1a)17. DNA barcoding has been used to track cell lineages during experimental evolution18, neuron dispersal19, stem cell differentiation20 or the development of drug resistance in cancer cells21. Furthermore, DNA barcodes chromosomally integrated at strain-specific neutral sites using, for example, homologous recombination or RNA-guided nucleases such as CRISPR–Cas9 are used to study host bottlenecks and subsequent tissue-specific population dynamics of bacterial pathogens in animal systems22,23,24. In plant–microbe interactions, DNA barcodes have been used mainly for screening bacterial mutant libraries based on a method called BarSeq25,26,27 and to distinguish closely related Pseudomonas strains in the phyllosphere of Arabidopsis thaliana28. However, simultaneous profiling of taxonomically diverse microbial communities by amplicon sequencing of marker genes and of DNA barcodes of near-isogenic strains is not established in the field of vertebrate or plant microbiota studies.
Here we describe a modular bacterial tag (MoBacTag) tool to label a broad range of taxonomically distinct bacteria with a DNA barcode as well as a fluorescent tag. The DNA barcode allows discrimination of bacteria that cannot be distinguished when profiling microbial communities using conventional marker gene amplicon sequencing. As the DNA barcode is flanked by primer binding sites specific to V5–V7 16S rRNA, fungal ITS, plant ITS and barcodes, the abundances of barcoded bacteria can be determined simultaneously with abundances of unlabelled bacteria and fungi by amplicon sequencing of the corresponding marker genes. Furthermore, DNA-barcode-harbouring plasmids are used as spike DNA to estimate microbial load and calculate the ratio of plants to microbes. As a proof of principle, we use the MoBacTags to recapitulate the colonization defect of Pseudomonas capeferrum WCS358 cyoB and pqqF immunosuppressive mutants29. By simultaneously analysing DNA barcodes and 16S rRNA sequences, we reveal an activity specific to the WCS358 pqqF mutant in community establishment, which is not trans-complemented by wild-type WCS358.
Results
Design of MoBacTag tools
MoBacTag tools have been designed for orientation-specific high-frequency insertion into bacterial chromosomes at the conserved Tn7 attachment site30 downstream of the glmS gene that is present in 98.6% (426 out of 432) of all bacterial draft genomes in the Arabidopsis-derived bacterial culture collection (At-R-SPHERE31; Extended Data Fig. 1)32. To show broad utility, we have labelled a total of 22 plant-derived bacterial strains from 9 different genera with MoBacTags by Tn7-mediated chromosomal integration (Achromobacter, Deinococcales, Lysobacter, Neorhizobium, Pseudomonas, Pseudoxanthomonas, Rhizobium, Rhodanobacter, Xanthomonas; Supplementary Table 1).
The chromosomally integrated fragment contains (1) the minimal Tn7 elements (Tn7R, Tn7L)33, (2) two terminator sequences, (3) an antibiotic marker flanked by yeast Flp recombinase recognition sites for sequential excision, (4) a fluorescent marker and (5) a barcode DNA tag (Fig. 1b). Regulatory elements, expression cassettes and the barcode DNA tag were first individually mobilized into level 1 vectors using modular cloning principles (Supplementary Table 3)34. All 20 possible combinations of the four antibiotics and five fluorescent markers were assembled into a modular cloning-adapted pSEVA211-based backbone35. For each antibiotic–fluorescent marker combination, we constructed three to five ready-to-use vectors with distinguishable barcode DNA tags (Supplementary Table 5). Chromosomal integration into attTn7 is enforced by the pSEVA211 vector backbone containing the restrictive R6K origin of replication36, which renders the vector unstable in most bacteria, and the negative selection marker sacB (Extended Data Fig. 2).
For profiling of microbial communities by amplicon sequencing, the barcode DNA tag is flanked by conserved bacterial V5–V7 16S rRNA sequences, conserved fungal ITS1 and ITS2 sequences, plant ITS-p4 and ITS-p5 sequences37,38, and barcode-specific primer binding sites (Fig. 1b). Unique barcode DNA tags were generated by random blunt-end ligation of an equimolar ratio of ten different, double-stranded oligonucleotides (38 nucleotides (nts)), each consisting of four pyrosequencing-friendly barcodes, followed by DNA fragment size selection (Fig. 1c). The final barcode DNA tags preferentially consisted of an array of four oligonucleotides, resulting in an average length of approximately 150 nts per barcode (Fig. 1c). Preassembled barcode DNA tags were then integrated in exchange for a negative ccdB selection cassette previously integrated between primer binding sites of recipient vectors (Fig. 1d).
MoBacTag plasmid DNA was additionally used as spike-in DNA during library preparation for amplicon sequencing and comparative analysis (Fig. 1b and Supplementary Information). As the 16S, fungal ITS and plant ITS primer binding sites flank the barcode DNA tag, read counts assigned to chromosomally integrated barcode DNA tags, bacterial 16S rRNA or fungal ITS can be normalized to read counts specific to spike-in barcode DNA tags. These ratios can then be used to calculate bacterial-to-plant, fungal-to-plant or bacterial-to-fungal ratios. Finally, these ratios provide an estimate of the microbial load of the corresponding microbial kingdom in the sampled plant compartment (Figs. 1a and 2a).
a, Principles of relative abundances and normalization using a spike plasmid in bacterial profiling. b,c, Linear correlation of normalized spike-specific read counts with spike concentrations obtained from amplicon sequencing of root (n = 15) and peat (n = 15) matrix samples using bacterial-16S-specific (799, 1,192; b) and plant-ITS-specific (ITS-p4, ITS-p5; c) primers. d, Linear correlation of normalized barcode-specific read counts with normalized 16S-specific read counts for correction factor adjustment of individual chromosomally integrated barcodes into WCS358 (n = 16 per variant, corresponding to n = 10 from root, n = 3 from peat matrix and n = 3 from input). e, Linear correlation of normalized barcode-specific read counts with normalized 16S-specific read counts for correction factor adjustment of individual chromosomally integrated barcodes into Rhizobiales R13D (n = 3 per variant). f, Spike-normalized uncorrected and correction-factor-adjusted abundance of read counts for barcode and 16S rRNA sequences within samples with increasing numbers of equimolar mixed Rhizobiales R13D barcode strains. The colours indicate the DNA barcode tags.
Validation of tags as spike and artificial 16S rRNA sequence
Correlation of spike read counts with the spike DNA concentration was tested for each oligonucleotide pair in root and peat matrix samples to ensure that spike DNA abundance was reflected in read counts specific to barcode DNA tag (Fig. 2a–c and Extended Data Fig. 3a,b). We first normalized all read counts using spike 1 reads to compensate for different sequencing depths. Normalized spike-2-specific barcode read counts from amplicon sequencing with primers specific to 16S rRNA, fungal ITS, plant ITS or the barcode correlated linearly over five orders of magnitude with spike 2 plasmid DNA concentrations in both root and peat matrix samples.
We compared the efficiencies of PCR amplification and DNA sequencing for natural 16S rRNA with corresponding barcode DNA tags to test for previously reported PCR biases in microbiota profiling using 16S rRNA sequences39. To exclude possible biases due to known variations in 16S rRNA copy numbers in different bacterial taxa40, different MoBacTags (termed tag119, tag120 and tag190) were inserted into wild-type P. capeferrum WCS358, and the WCS358:cyoB and WCS358:pqqF mini-Tn5 transposon insertion mutants29, at identical Tn7 chromosomal integration sites. Short-read sequencing of individual MoBacTag-labelled bacteria revealed 3.5-fold higher read counts for tag119, 1.9-fold higher for tag120 and 1.8-fold higher for tag190 compared with the corresponding natural endogenous 16S rRNA read counts of P. capeferrum WCS358 (Fig. 2d). These data indicate that despite identical chromosomal integration sites, primer binding sites and 16S rRNA genetic backgrounds, individual barcode DNA tags exhibit different tag-to-16S read count ratios regardless of primers used for amplification (Fig. 2, Extended Data Fig. 3c,d and Supplementary Information). To next test whether the tag-specific amplification rates are unique to the Tn7 locus, we also generated mutants of the root commensal Rhizobiales R13D by integrating MoBacTags (termed tag104, tag93, tag94, tag91) into different loci through homologous recombination. The data also showed tag-specific amplification with read counts that were 1.6-fold higher for tag104, 1.5-fold higher for tag93 and 1.4-fold higher for tag94 compared with the corresponding natural endogenous 16S rRNA read counts (Fig. 2e). Fewer read counts were obtained for DNA tag91 than for endogenous 16S rRNA (Fig. 2e), indicating tag-specific amplification bias for MoBacTags inserted at different, but also identical, chromosomal loci. To ensure that the read counts obtained from different barcode DNA tags accurately reflect the respective bacterial abundance, the tag-to-16S count ratios were used as a correction factor for the read counts specific to the barcode DNA tag. The corrected tag-specific read counts matched the read counts specific to 16S rRNA obtained from the sequencing of pure bacterial cultures (Fig. 2f). Thus, correction factors need to be determined for each MoBacTag-labelled bacterial strain. We then investigated potential combinatorial effects of multiple MoBacTags within a sample. Genomic DNA of R13D strains carrying the four different barcode DNA tags were mixed in a 1:1 ratio with increasing complexity, that is, two, three or all four strains (Fig. 2f). Cumulative tag-specific read counts were comparable to 16S rRNA read counts only after applying tag-specific correction factors irrespective of sample complexity, that is, two, three or four barcode DNA tags. Similar read counts were obtained for each tag per condition when using the tag-specific correction factors determined from the ratios of tag to 16S reads. In summary, the number of MoBacTag-labelled strains within a condition does not change the number of tag-specific reads per strain, so the corrected tag-specific reads reflect strain abundance.
MoBacTag labelling of diverse root microbiota members
As with any Tn7-mediated insertion, genome integration of the MoBacTag into the bacterial strain of interest requires four (not necessarily consecutive) working days, assuming that a DNA transformation protocol has already been established for the bacterium of interest (Fig. 3a)33. For conjugation-based tagging of root-derived Rhodanobacter R179, Tn7 attachment sites and natural antibiotic resistances were investigated. For conjugation, MoBacTag multigene vectors were mobilized into auxotrophic, conjugation-competent Escherichia coli BW29427 (ref. 41). In addition to selecting transformants based on MoBacTag-encoded antibiotic resistance, selection against antibiotic-resistant E. coli BW29427 was achieved by depletion of diaminopimelic acid (DAP), which is required for the survival of E. coli BW29427 but not the target strains42. The presence of the MoBacTag fragment was verified by PCR of bacterial colonies using plant-ITS-specific oligonucleotides (Fig. 3b; JT103 and JT108). The absence of PCR-amplified fragments from non-transformed bacterial colonies confirmed the specific binding of the plant ITS oligonucleotides to the MoBacTag in Rhodanobacter R179. In community profiling experiments with 15 bacterial strains selected from the At-R-SPHERE collection (synthetic communities (SynCom) modified from a previous study43), no bacteria-derived sequences were recovered using the plant-ITS-specific oligonucleotides, supporting the target specificity of the plant ITS primers. In addition, transformants were genotyped with PCR using oligonucleotides specific to 16S rRNA, resulting in two size-separable PCR products, as the barcode DNA tag is shorter than the endogenous 16S rRNA amplicon (Fig. 3b; oligonucleotides 799 and 1192). Chromosomal integration was validated using a combination of Tn7-specific and glmS-strain-specific oligonucleotides (Fig. 3b)33. Further PacBio long-read genome re-sequencing of MoBacTag-labelled and wild-type Arabidopsis-root-derived Rhizobiales strains revealed no chromosomal rearrangements induced by the Tn7-mediated insertion (Extended Data Fig. 4a). Thus, the tagged strains differ from the wild type only by the chromosomally integrated MoBacTag, which could be validated by PCR-based genotyping. Comparisons of unlabelled and MoBacTag-labelled strains showed indistinguishable colony morphologies and growth curves in liquid medium (Extended Data Fig. 4b). Moreover, the composition of a microbial community was largely unaltered by the presence of a MoBacTag chromosomally integrated into the P. capeferrum WCS358 plant microbiota member (Extended Data Figs. 5 and 6 and Supplementary Information).
a, General timeline for labelling a target strain by chromosomal MoBacTag integration. Depending on the target strain, Tn7-mediated insertion requires four (not necessarily consecutive) working days. b, Validation of MoBacTag transformation and chromosomal integration. DNA was extracted from transformed (+BC) and non-transformed Rhodanobacter R179 strains, PCRs were performed using indicated primers and amplicons were visualized on an agarose gel. To test barcode integration, Tn7-element- and glmS-specific primers were used. This experiment was performed two times independently, and similar results were obtained. c, MoBacTag-labelled strains express fluorescent proteins. Expression of MoBacTag-encoded fluorescent protein was detected using live confocal laser scanning microscopy in liquid medium (four panels on the left; observed two times independently) and on A. thaliana roots (panel on the right; observed five times independently). Representative images are shown. Scale of bacteria in liquid culture (left), 10 µm; scale on roots (right), 20 µm.
MoBacTags encoding different fluorescent markers were transformed into the root-derived commensal Rhodanobacter R179. Live cell imaging using confocal laser scanning microscopy was used to detect the expression of the four chromosomally encoded fluorescent markers driven by the tac promoter in bacteria cultivated on nutrient-rich medium or in association with the plant host (Fig. 3c). Expression of fluorescent proteins was also detected in MoBacTag-labelled P. capeferrum WCS358, Pseudomonas simiae WCS417, Xanthomonas campestris pv. vesicatoria and Rhizobium R13D, indicating a robust activity of the tac promoter in the corresponding bacterial taxa (Extended Data Fig. 4c).
Spike normalization to determine microbial load in planta
To investigate whether the immunosuppressive activity of P. capeferrum WCS358 selectively promotes its own colonization or also influences root colonization by other members of the bacterial community, wild-type WCS358 and the immunosuppressive mutants WCS358:pqqF and WCS458:cyoB29 were each, or in combination, co-inoculated with a taxonomically diverse SynCom consisting of 15 Arabidopsis-root-derived bacteria from the At-R-SPHERE culture collection on germ-free Arabidopsis seedlings43,44. The corresponding wild-type genes contribute to the production of gluconic acid and its derivative 2-keto gluconic acid, which are proposed to suppress plant immunity by lowering extracellular pH locally29. To track individual strains in the SynCom, wild-type WCS358, WCS358:pqqF and WCS358:cyoB were labelled with MoBacTags differing in DNA barcodes, but an identical antibiotic resistance cassette, so that potential marginal effects of the MoBacTag on community establishment were identical for all conditions (Extended Data Fig. 5).
We first focused on the SynCom members by in silico depletion of WCS358 16S rRNA reads. Read counts relative to sample read depth indicated a higher relative abundance of Streptomyces spp. R1310 and a decreased relative abundance of Microbacterium spp. R61 in the presence of the WCS358:pqqF mutant compared with the WCS358 wild type, which is not indicative of altered absolute abundances (Fig. 4a). Normalization of 16S rRNA reads to spike or plant-ITS-derived reads revealed a substantially reduced total bacterial load on roots colonized by the WCS358:pqqF mutant (Fig. 4b,c). Therefore, the altered relative abundances of Streptomyces spp. R1310 and Microbacterium spp. R61 in the presence of WCS358:pqqF might incorrectly hint at absolute changes due to the compositional nature of community profiling in the absence of normalization. As a result, further analyses were performed only on the spike-normalized data.
a–c, Relative (a), spike-normalized (b) and plant-ITS-normalized (c) abundance of SynCom members co-inoculated with P. capeferrum WCS358 wild type, and WCS358:pqqF or WCS358:cyoB mutant on A. thaliana roots, plotted as stacked bar plots. Colours indicate SynCom members. The mean of 12 biological samples collected from three independent replicates is shown. d,e, Constrained coordination of the microbial profile of a 15-member SynCom in the presence of either P. capeferrum WCS358 wild type (dark grey, n = 12), WCS358:pqqF (salmon, n = 12) or WCS358:cyoB (light green, n = 12) mutant (d), or WCS358 wild type plus WCS358:pqqF (brown, n = 12) or additional WCS358:cyoB (light orange, n = 12) on A. thaliana roots (e). The n values indicate biological samples collected from three independent replicates. The P values indicate statistical significance determined by permutational analysis of variance test between the compositions of SynCom 1 and SynCom 2 co-established with WCS358 derivatives indicated by the coloured boxes (permutation = 999). Ellipses correspond to Gaussian distributions fitted to each cluster (95% confidence interval). Shapes represent independent experiments. wt, wild type; CPCoA, constrained principal coordinate analysis. f,g, The normalized abundance of strains in the root compartment on co-inoculation with P. capeferrum WCS358 wild type (dark grey, n = 12), and the pqqF (salmon, n = 12) or cyoB (light green, n = 12) mutant: highly abundant (f) and low abundant (g) strains. The n values indicate biological samples collected from three independent replicates. The P values indicate statistical significance determined between the wild type and the mutants using a two-sided Dunn’s test. The box plots centre on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the 1.5× interquartile range.
Constrained principal coordinate analysis of Bray–Curtis dissimilarities of spike-normalized 16S rRNA reads showed that the reduced SynCom load on roots in the presence of the WCS358:pqqF mutant compared with wild-type WCS358 correlates with the formation of distinct communities (Fig. 4d,e). Seven SynCom members showed a significantly reduced root colonization capacity in the presence of WCS358:pqqF compared with the WCS358 wild type (Fig. 4f,g). The pqqF gene product is essential for the biosynthesis of pyrroloquinoline quinone (PQQ)45, which serves as a redox-sensitive co-factor of several bacterial dehydrogenases including the glucose dehydrogenase required for the production of gluconic acid and its derivative46. The reduced total microbial load can be explained by a similar trend for half of all SynCom members, suggesting that PQQ biosynthesis and not gluconic acid production by wild-type WCS358 promotes root colonization by taxonomically diverse members of the root microbiota. Unexpectedly, however, root-associated communities established in the presence of wild-type WCS358 plus WCS358:pqqF showed only a slight shift towards communities containing wild-type WCS358 alone (Fig. 4b,c,e). Thus, the presence of the PQQ-deficient WCS358:pqqF strain has an unexpected dominant influence on root microbiota establishment that is not complemented by co-inoculation with the PQQ-producing wild-type WCS358. This suggests that immunosuppression mediated by gluconic acid synthesis in wild-type WCS358 is insufficient to support the establishment of wild-type-like root communities in the presence of the WCS358:pqqF mutant. We hypothesize that this lack of trans-complementation by wild-type WCS358 might be related to PQQ consumption owing to proliferation of the WCS358:pqqF mutant, reducing the PQQ pool available for the SynCom. We tested whether WCS358 can import extracellular PQQ. Excess of d-glucose mediates growth inhibition through medium acidification by the activity of PQQ-dependent glucose dehydrogenase (GDH46). Consistent with this, the growth of the WCS358:pqqF mutant in vitro was not restricted by an excess of d-glucose compared with the growth of wild-type WCS358 (Extended Data Fig. 7). Chemical PQQ supplementation of the WCS358:pqqF mutant restored wild-type-like d-glucose-dependent growth restriction, suggesting that WCS358 imports PQQ.
Unlike WCS358:pqqF, the loss of cyoB in WCS358 had only a minor effect on the total bacterial load and the composition of the root-associated communities (Fig. 4d,f,g). As genetic depletion of cyoB in P. capeferrum WCS358 eliminates 2-keto-d-gluconic acid production but preserves residual gluconic acid biosynthesis in vitro29, the wild-type-like community found here in the presence of WCS358:cyoB might be explained by residual gluconic acid production in planta.
Tracking of near-isogenic strains in microbiota
Next, we asked whether the reduced root colonization by the WCS358:cyoB and WCS358:ppqF mutants reported from previous inoculation experiments in an unsterilized soil–sand matrix29 can be reproduced in a peat-matrix-based gnotobiotic plant system47 in the presence of a defined 15-member bacterial community. Consistent with the relative 16S rRNA read counts for the 15 members of the SynCom (Fig. 4a), the interpretation of the relative 16S rRNA read counts specific to P. capeferrum WCS358 was limited by the differential total microbial load between conditions (Fig. 5a). Accordingly, no difference in abundance was detected between the wild-type WCS358 and the WCS358:pqqF mutant when analysing the relative read counts. However, the spike-normalized abundances of 16S rRNA reads from P. capeferrum WCS358 strains derived from either wild type, or WCS358:cyoB or WCS358:pqqF mutants, recapitulated the reduced root colonization by the mutants in our peat-based gnotobiotic plant system previously observed in an unsterilized soil–sand matrix (Fig. 5b). In agreement with published data, the abundance of wild-type WCS358 and WCS358 mutants in the peat matrix compartment was comparable (Fig. 5a,b)29. Thus, pqqF and cyoB are needed to specifically promote WCS358 colonization of the root compartment but are dispensable for bacterial growth in the peat matrix.
a,b, Relative (a) and spike-normalized (b) abundance of P. capeferrum WCS358 wild type, WCS358:cyoB and WCS358:pqqF in the root (n = 36) and peat matrix (n = 14) compartment quantified by 16S rRNA read counts. The n values indicate biological samples collected from three independent replicates. The letters indicate statistical significance (P < 0.05) determined using a two-sided Dunn’s test. The bar plots represent the mean ± s.e.m. c,d, The normalized abundance calculated using barcode-specific reads of P. capeferrum WCS358 wild type, and the pqqF or cyoB strains in the root (n = 60) (c) and peat matrix (n = 31) (d) compartment on co-inoculation with the 15-member SynCom alone, or in indicated mixtures. The n values indicate biological samples collected from three independent replicates. The letters indicate statistical significance (P < 0.05) determined using a two-sided Dunn’s test. The box plots centre on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the 1.5× interquartile range. NS, not significant.
To test whether strain-resolved abundances of MoBacTags can be retrieved during community profiling of bacteria with identical 16S rRNA sequences, the abundances of the MoBacTags incorporated into P. capeferrum WCS358 wild type, and the WCS358:pqqF and WCS358:cyoB mutants, were examined. Spike-normalized, corrected barcode DNA tag reads independently confirmed similarly reduced root and wild-type-like peat matrix colonization by the tested WCS358 mutants when only one WCS358 genotype was added to the 15-member SynCom at a time (Fig. 5c,d). Interestingly, co-inoculation with wild-type WCS358 did not elevate either WCS358:pqqF or WCS358:cyoB abundance in the root compartment (Fig. 5c). Thus, the WCS358 wild type did not trans-complement for the impaired root colonization of WCS358 mutants. On the contrary, in these mixed inoculation experiments (wild-type WCS358 plus WCS358 mutants), the abundance of wild-type WCS358 and to a greater extent that of WCS358:cyoB on roots decreased upon co-inoculation with WCS358:pqqF (Fig. 5c), as was the case for most other SynCom members (Fig. 4f,g). Furthermore, after co-inoculation of both WCS358 mutants, wild-type WCS358 and the 15-member SynCom, we detected fewer than 10 reads for WCS358:cyoB in all 12 root and most (8 out of 11) matrix samples. This corresponds to a >100-fold reduction compared with wild-type WCS358 reads, indicating that WCS358:cyoB was essentially outcompeted in this condition (Fig. 5d). The tag-based strain abundances reveal previously unsuspected additional and distinct functions of pqqF compared with cyoB on roots. In summary, the MoBacTag allowed us to test for complementation or competition between strains encoding identical 16S rRNA sequences during root microbiota establishment.
To show broad utility of MoBacTags, we inoculated MoBacTag-labelled strains Rhizobium R13C and R13D in two natural soils, each containing a highly complex resident microbial community, and in a Jiffy peat matrix (Extended Data Fig. 8a–d). Five weeks after cultivation of Arabidopsis Col-0 in these substrates, reads specific to the barcode DNA tags of R13C and R13D were detected in samples from unplanted control substrates, planted substrates and root compartments (Extended Data Fig. 8e–g). Thus, MoBacTag can also be used to track near-isogenic bacterial strains in highly complex resident communities of natural soils.
Discussion
We have developed and validated DNA barcodes, which are co-amplified with natural endogenous bacterial V5–V7 rRNA, fungal ITS or plant ITS sequences by integrating respective primer binding sites into the DNA barcode. The MoBacTag enables direct tracking of multiple near-isogenic strains during conventional bacterial 16S rRNA or fungal ITS community profiling. Since MoBacTag plasmids are based on modular cloning principles, each position within the multi-gene construct can be customized34 by, for example, replacing Tn7 elements with genomic regions for homologous recombination or inserting (1) additional community profiling primer binding sites into the DNA barcode, (2) an expression cassette for complementation approaches or (3) available modular cloning-compatible elements such as intensity-optimized fluorescent markers48,49.
Bacterial DNA barcoding was applied to investigate antagonistic interactions of pseudomonad leaf strains co-occurring on Arabidopsis28. However, the DNA barcode architecture used does not allow co-amplification with endogenous marker genes, meaning that community analysis is restricted to barcoded strains only. The MoBacTag barcodes were semi-randomly assembled by random ligation of synthesized oligonucleotides consisting of 454 barcodes to avoid homopolymer or dipolymer tracts (Fig. 1). Unexpected PCR biases of individual 454-based barcodes required correction factors to determine the abundance of MoBacTag-labelled strains (Fig. 2d,e). As the error-corrected MoBacTags are chromosomally integrated as a single copy in most bacteria, tag-based abundance estimates can avoid biases due to variations in 16S rRNA gene copy number from 1 to 15 between bacterial species40,50. Our spike-in plasmid architecture is similar to the architecture of other spike-in plasmids that require quantitative PCR measurements (Fig. 1a)51,52. Our spike plasmid allows for direct 16S rRNA read count normalization and normalization to plant ITS read counts. The latter enables studying the composition and load of microbial communities in experiments with multiple plant species. However, differences in plant ITS copy number between plant species and accessions should be accounted for using correction factors. Besides spike-in normalization, host-associated microbe PCR (hamPCR)-based approaches are also used to measure microbial load and community composition53. MoBacTag and hamPCR approaches are complementary and could be combined to further increase the accuracy of host microbial load estimates.
The spike-in normalization revealed a significantly reduced SynCom load in the presence of the WCS358:pqqF mutant, but not WCS358:cyoB, which can be explained by significantly decreased abundances of 7 of the 15 SynCom members, each representing different core taxonomic lineages of the A. thaliana root microbiota (Fig. 4b,f,g)43,44. WCS358:pqqF and WCS358:cyoB are both impaired in the acidification of the extracellular space, although the WCS358:cyoB mutant produces residual amounts of gluconic acid29. Despite shared functions in the biosynthesis of gluconic acid, the WCS358:pqqF mutant, and not the WCS358:cyoB mutant, reduced microbial load on roots, which is not trans-complemented by gluconic-acid-producing wild-type WCS358. Thus, the observed reduction in microbial load is unlikely to be a direct consequence of impaired gluconic-acid-mediated host immunosuppression. We propose instead that the co-factor PQQ serves as a common good within the root-associated bacterial community, as has been shown for other cofactors such as cobamides in bacterial co-cultures composed of different species54. Extracellular PQQ levels are probably relevant in bacterial communities, as PQQ import has been shown to be concentration dependent: the co-factor is imported by diffusion at high concentrations, whereas active TonB-dependent import is required at low concentrations, at least for E. coli55. Thus, the WCS358:pqqF mutant could reduce the extracellular PQQ pool on roots owing to lack of PQQ biosynthesis despite continuous PQQ consumption through WCS358:pqqF proliferation, resulting in PQQ deficiency in the bacterial community. This would also explain why WCS358:pqqF-mutant-specific community phenotypes are not trans-complemented by wild-type P. capeferrum WCS358 in planta.
MoBacTag DNA barcodes generated in vitro were designed for tracking near-isogenic bacterial strains in community contexts across multiple generations. These strains cannot be distinguished by endogenous barcodes such as the V5–V7 16S rRNA region. The presence of the dominant-negative ccdB selection marker in the recipient vector (Fig. 1) should enable use of MoBacTags for pool transformation approaches, each with randomly loaded DNA barcode tags (Fig. 1)17. This opens up future opportunities for MoBacTag-based lineage tracking during experimental evolution of microbial communities56, as potential compensatory growth responses of community members to changes in labelled lineages can be tracked in parallel. Addition of primer binding sites for other eukaryotic hosts will increase the versatility of MoBacTag vectors for studies of any host–microbiota interaction.
Methods
Bacterial media and growth conditions
Level 1 vectors were transformed into E. coli DH5 and DB3.1 for ccdB-encoding plasmids (Supplementary Table 1). Level 2 pBCC and pBC vectors were transformed into E. coli DB3.1λpir57 and BW29427, also known as WM306441, respectively (Supplementary Table 1). pTNS3 (ref. 58) was also cloned into E. coli BW29427. Transformation of the R6K origin of replication encoded on MoBacTags into bacteria with methyl-specific restriction systems was enabled by chromosomal integration of pir2, which is responsible for R6K-based replication, into a non-methylating conjugation-competent E. coli (ET12567/pUZ8002)59,60. Therefore, pir2 was transformed into E. coli ET12567/pUZ8002 via Tn7-mediated chromosomal integration similar to the procedure of MoBacTag labelling. pir2 was first mobilized into pUC18-miniTn7-Gent-GW61 using Gateway cloning according to the user manual.
E. coli were cultivated in Luria–Bertani medium (25 g l−1 Luria–Bertani medium; Sigma) at 37 °C, and commensal bacterial strains were cultivated in 0.5 tryptic soy broth (TSB, 15 g l−1; Sigma) or TY medium (5 g l−1 tryptone, 3 g l−1 yeast extract, 10 mM CaCl2) at 25 °C. Media were supplemented with 15 g l−1 Bacto Agar (Difco) for solidification. Antibiotics or DAP were added to the media, when necessary, at the following concentrations: streptomycin (Sm, 100 μg ml−1), spectinomycin (Sp, 100 μg ml−1), tetracycline (Tc, 10 μg ml−1), gentamicin (Gm, 25 μg ml−1), kanamycin (Kn, 50 μg ml−1) and DAP (50 μg ml−1).
Assembly of unique DNA barcodes by random ligation of oligonucleotides
Complementary oligonucleotides (Supplementary Table 2) were mixed at an equimolar ratio (10 µM), incubated at 94 °C for 2 min and gradually cooled. Double-stranded oligonucleotides were then mixed in a ratio of 1:1 and prepared for blunt-end ligation using the End-It DNA End-Repair Kit according to the user manual (Biosearch Technologies). Ligation was performed overnight at 4 °C using T4 DNA ligase (New England Biolabs) followed by heat inactivation for 10 min at 80 °C. The preassembled barcodes were finally selected for 200–300 bp fragments by BluePippin from Sage Science at the Max Planck Genome Centre, Cologne, Germany (https://mpgc.mpipz.mpg.de/home/). Size-selected arrays of ligated oligonucleotides were then cloned between primer binding sites of pBCC vectors (see next paragraph).
Generation of MoBacTag plasmids using the modular cloning strategy
Expression cassettes, terminator sequences and mini-Tn7 elements were amplified using PrimerStar Max DNA Polymerase from Takara (templates indicated in Supplementary Table 3). PCR fragments were cloned into level 1 recipient plasmids (Supplementary Table 3) from the MoClo Toolkit by a restriction and ligation reaction with BsaI and T4 DNA ligase (New England Biolabs) using ligase buffer34,62. BsaI, BpiI and SmaI recognition sites were removed by altering a single nucleotide, without changing the encoded amino acid, in the recognition site using primers with restriction-site-mutating sequences (Supplementary Tables 2 and 3). The ccdB cassette was flanked with SmaI recognition sites and amplicon sequencing primer binding sites by consecutive PCRs with tailed primers (Supplementary Table 2) and then cloned into the level 1 recipient plasmid plCH47761. The antibiotic markers (pBCC030, pBCC031, pBCC032) were flanked with Flp recombinase target (FRT) sites by two consecutive PCRs using FRT site-encoding tailed primers, whereas for pBCC033, the FRT sites were already present in the donor plasmid (Supplementary Table 3). First, a green fluorescent protein (GFP)-encoding plasmid was assembled (pBCC029) by amplifying GFP from pUC18-mini-Tn7T-Tp-gfpmut3 and pTAC from pLM449 with tailed primers integrating BsaI restriction sites, and this was cloned into the plCH47751 level 1 receptor by a restriction and ligation reaction with BsaI and T4 DNA ligase using ligase buffer. Finally, the pTAC promoter and coding sequence of the fluorescent markers were amplified from pLM426 derivatives (Supplementary Table 3) and combined with the pBCC0029 recipient plasmid including a terminator sequence by In-Fusion HD from Takara. All plasmids for this study were purified using the NucleoSpin Plasmid kit (Macherey-Nagel). Level 1 domesticated sequences were then assembled into a pSEVA211-based35 level 2 recipient vector by a BpiI restriction and ligation reaction, resulting in BarCode Construction (pBCC) multi-gene constructs (Supplementary Table 3). To this end, a red fluorescent protein (tagRFP) expression cassette was amplified by PCR integrating BpiI recognition sites and cloned into the pSEVA211 by a restriction and ligation reaction with BsaI, EcoRI, HindIII and T4 DNA ligase (New England Biolabs). Preassembled barcodes were inserted by SmaI restriction and ligation as follows: 45 cycles of 5 min at 16 °C, and 5 min at 30 °C followed by 5 min 95 °C. Then, fresh SmaI was added and incubated at 30 °C for 30 min resulting in final MoBacTag vectors. Barcode tag sequences were identified for individual E. coli BW29427 colonies from pooled transformation with MoBacTag vectors by colony PCR with primers MobacTag barcode F/R (Supplementary Table 5), followed by Sanger sequencing (Eurofins Scientific). Sequencing results were analysed in CLC Main Workbench (QIAGEN). The plasmids generated for the MoBacTag kit will be made available via Addgene.
Labelling of bacterial strains with MoBacTags using the mini-Tn7 system
MoBacTag vectors were transferred into commensal bacterial strains (Supplementary Table 1) by triparental mating. Saturated liquid cultures of the recipient commensal strain, E. coli strain BW29427/pTNS3 and BW29427/pBC were mixed in a 1:2:2 ratio and incubated for 24–48 h at 25 °C. Afterwards, transformants were selected on 0.5 TSB or TY medium containing 10% sucrose and the corresponding antibiotics. Bacterial DNA was extracted by re-suspending a bacterial colony in 25 µl buffer I (25 mM NaOH, 0.2 mM EDTA, pH 12) followed by incubation at 95 °C for 30 min and addition of 25 µl buffer II (40 mM Tris–HCl). The genomic insertion of the MoBacTag was validated by PCR as described in a previous study33.
AttTn7 box conservation across the At-R-SPHERE culture collection
The glmS sequences were extracted from the whole-genome assemblies of every strain included in the At-SHPERE culture collection44. The last 12 amino acids or the last 36 DNA bases from the extracted glmS sequences were aligned and visualized with the software WebLogo (https://weblogo.berkeley.edu/logo.cgi).
Bacterial genome assembly
The Max Planck Genome Centre, Cologne, Germany (https://mpgc.mpipz.mpg.de/home/), performed the DNA isolation of wild-type and MoBacTag-labelled strains and also the sequencing on the Pacific Biosciences Sequel IIe platform. Reads were assembled de novo using the Hifiasm software (https://github.com/chhylp123/hifiasm). To compare wild-type and MoBacTag-labelled strains, we used Mauve software for reordering the contigs (https://darlinglab.org/mauve). Genomes were compared by generating a dot plot on genome scale using the Genome Pair Rapid Dotter (gepard) software (https://doi.org/10.1093/bioinformatics/btm039).
MoBacTag-labelled and unlabelled Rhodanobacter growth in liquid monoculture
MoBacTag-labelled and unlabelled Rhodanobacter R179 strains were grown in six replicates each in 0.5 TSB as individual cultures in a 96-well bacterial culture plate (Greiner-CELLSTAR 96-well plate, transparent, flat bottom; Sigma-Aldrich) at 25 °C. Absorbance was measured every 10 min, 10 s after 20 s of shaking (290 rpm), using a microplate reader (Infinite M200 PRO, Tecan) at 600 nm. The mean values of four measurements per well were used for the analyses.
PQQ- and glucose-dependent P. capeferrum growth in liquid monoculture
P. capeferrum wild-type WCS358 and the WCS358:pqqF mutant were grown in four replicates each in modified XVM2 minimal medium (20 mM NaCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl2, 0.01 mM FeSO4, 10 mM succinate, 0.03% Casamino acids) supplemented with 110 mM d-glucose and 3 µM PQQ as individual cultures in a 96-well bacterial culture plate (Greiner-CELLSTAR 96-well plate, transparent, flat bottom; Sigma-Aldrich) at 25 °C. Absorbance was measured every 10 min, 10 s after 20 s of shaking (290 rpm), using a microplate reader (Infinite M200 PRO, Tecan) at 600 nm. The mean values of four measurements per well were used for the analyses.
Visualization of fluorescently labelled commensal bacteria
MoBacTag-labelled bacterial strains were harvested from 0.5 TSB plates and re-suspended in 10 mM MgSO4. The root colonization assay was performed as follows: surface-sterilized A. thaliana Col-0 seeds were sown on agar plates (1% Bacto agar, BD Biosciences) prepared with MS/2 (as described for the Gnoptopot system) and supplemented with MoBacTag-labelled Rhodanobacter R179 or Rhizobiales R13D at a final concentration of OD600 = 0.0005 (ref. 63). Washed bacteria (‘Bacterial microbiota reconstitution experiment’ in Methods) were mixed into the medium before solidification. After 14 days of growth under short-day conditions (10 h light, 21 °C; 14 h dark, 19 °C), bacteria were visualized on roots. Confocal laser scanning microscopy was performed on a Zeiss LSM880 inverted confocal scanning microscope. Pictures were taken with an LD C-Apochromat 40×/1.1 water immersion objective. To image root colonization, Z-stacks were generated, and maximum intensity projections were compiled. The following excitation and detection windows were used: GFP: 488 nm, 493–598 nm; cyan fluorescent protein: 458 nm, 472–528 nm; tagRFP: 561 nm, 582–754 nm; and far-red fluorescent protein: 561 nm, 591–759 nm.
Investigation of MoBacTag amplification biases by qPCR
For qPCR, genomic DNA extracted from WCS358, WCS358:pqqF and WCS358:cyoB as described in ‘Labelling of bacterial strains with MoBacTags using Mini-Tn7 system’ in Methods was used. Dilution series were used for qPCR using the iQ SYBR Green Supermix (Bio-Rad). Reactions (20 µl) were prepared with 3 μl genomic DNA, 10 μl SYBR Green Supermix and 0.4 μl of each primer (10 µM, Supplementary Table 6). qPCR was performed using the CFX ConnectTM Real-Time System (Bio-Rad) with the following conditions: 95 °C for 3 min; 95 °C for 15 s, 65 °C for 15 s and 72 °C for 15 s for 4 cycles; and 95 °C for 15 s, 57 °C for 15 s and 72 °C for 15 s for 39 cycles followed by a melting curve.
Bacterial microbiota reconstitution experiments
Saturated bacterial liquid cultures were pelleted by centrifugation at 8,000g for 5 min, followed by two washes with 10 mM MgSO4. Equivalent amounts of each strain were combined to yield the desired SynComs with an optical density (OD600) of 2. Aliquots of individual strains and the SynComs were taken and stored at −80 °C. The inoculum solution was prepared with MS/2 (2.22 g l−1 Murashige and Skoog basal salts, Sigma; 0.5 g l−1 MES anhydrous, BioChemica; adjusted to pH 5.7 with KOH) and the SynCom at a final OD600 of 0.02. We used the Gnotopot system47 to grow A. thaliana Col-0 plants with the bacterial SynComs (Supplementary Table 6). Each pot was inoculated with the bacterial SynCom by decanting 10 ml of the inoculum solution. With the use of a syringe, the excess liquid was removed from the box. A. thaliana seeds were surface sterilized by incubation in 70% ethanol twice for 5 min, followed by a brief wash with 100% ethanol. Seeds were then washed three times with sterile water and cold stratified for 2 days. Six sterilized seeds were placed on the matrix of each pot (Jiffy-7 pellets, Jiffy Products, https://www.jiffygroup.com/) and incubated under short-day conditions for 5 weeks (10 h light, 21 °C; 14 h dark, 19 °C). Roots were harvested by thoroughly removing attached soil using sterile water and forceps. Root and peat matrix samples were collected in Lysing Matrix E tubes (FastDNA Spin Kit for Soil, MP Biomedicals) and frozen in liquid nitrogen. Samples were stored at −80 °C until DNA isolation, which was performed using the FastDNA Spin Kit for Soil according to the user manual (MP Biomedicals).
Cultivation of MoBacTag-labelled Rhizobium strains in soil and Jiffy peat substrates
The MoBacTag-labelled Rhizobium R13C and R13D inoculum was prepared as above and inoculated in native Cas64 and Golm64 soil and non-autoclaved Jiffy peat (Jiffy-7 pellets, Jiffy Products, https://www.jiffygroup.com/) substrates to a final concentration (OD600 = 0.2 per strain). Eight surface-sterilized A. thaliana Col-0 seeds were sown on top and cultivated under short-day conditions for 5 weeks in a greenhouse (10 h light, 21 °C; 14 h dark, 19 °C). Pots were watered using deionized tap water throughout the experiment. Harvesting was performed as described for ‘Bacterial microbiota reconstitution experiments’ in Methods.
Bacterial community profiling by amplicon sequencing
Library preparation for Illumina MiSeq sequencing was performed as described previously5, except for the addition of 0.001 ng of pBCC023 or pBCC084 plasmid DNA per reaction to the master mix of PCR1, as spike. The final ratio of sample (6 ng) to spike (0.001 ng) DNA per PCR1 is 6,000. The oligonucleotides used for amplicon sequencing of 16S rRNA, fungal ITS and plant ITS sequences are listed in Supplementary Table 5. In all experiments, multiplexing of samples was performed by single or double indexing (only forward barcoded oligonucleotides for single indexing or forward and reverse barcoded oligonucleotides for double indexing). The indexed amplicon libraries were pooled, purified using Ampure (Beckman Coulter) and sequenced on the Illumina MiSeq platform.
To validate MoBacTag as a spike-in plasmid using the bacterial-16S-, fungal-ITS-, plant-ITS- and barcode-specific primers, 0.15 ng of pBCC069 plasmid was mixed with 5 ng of extracted DNA from root and peat matrix samples, prepared by Getzke et al.65. The standard curve was prepared with pBCC084 plasmid in tenfold dilution series from 1.5 ng to 0.00015 ng per PCR.
Processing of gene amplicon data
Amplicon sequencing data from SynCom experiments were de-multiplexed according to their barcode sequences and quality filtered using the USEARCH (v.10.0.240) and QIIME (v.2 2021.2) pipeline66. Paired-end reads were merged using the flash2 (v.2.2.00) software67. Quality-filtered merged paired-end reads were then aligned to reference amplicon sequences using Rbec (v.1.8.0)11. For the plant ITS sequencing data, only single-end reads were processed, because a sequence length of pITS (742 bp) cannot be merged from the Illumina 2 × 300 bp sequencing run. The reference sequences were extracted from whole-genome assemblies of every strain included in the SynCom, from the MoBacTag barcodes and from whole-genome assembly of A. thaliana Col-0 (TAIR9 assembly, www.arabidopsis.org). We checked that the fraction of unmapped reads did not substantially differ between compartments and experiments. Count tables were generated and used for downstream analyses of diversity in R (v.4.0.0) using the R package vegan68. Amplicon data were visualized using the ggplot2 R package69.
Normalized quantification of amplicon sequencing
Amplicon reads assigned to the spike were used to normalize plant ITS and bacterial 16S rRNA read counts similar to what was done in a previous study51. Identical amounts of spike-in plasmid DNA included for plant ITS and 16S rRNA library preparation were used for cross-normalization using the following equations:
Statistics and reproducibility
All experiments were performed in three full-factorial (biological and technical) replicates. Bacterial abundances were compared using an ANOVA test, followed by Tukey’s post hoc test (α = 0.05). Statistical tests on beta-diversity analyses were performed using a permutational analysis of variance test with 999 random permutations. Whenever box plots were used in figures, data were represented as median values (horizontal line), Q1 − 1.5 × interquartile range (boxes) and Q3 + 1.5 × interquartile range (whiskers).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw amplicon reads and genome assemblies have been deposited in the European Nucleotide Archive under the accession number PRJEB61076. For the A. thaliana Col-0 sequence, the TAIR9 assembly was used (www.arabidopsis.org). The bacterial At-R-SPHERE 16S sequences were obtained from the website https://www.at-sphere.com. Source data are provided with this paper.
Code availability
The scripts used for the computational analyses described in this study are available at https://github.com/thouinjulien/MoBacTag, to ensure replicability and reproducibility of these results.
References
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T. & Singh, B. K. Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621 (2020).
Castrillo, G. et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017).
Harbort, C. J. et al. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28, 825–837 (2020).
Berendsen, R. L., Pieterse, C. M. & Bakker, P. A. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012).
Duran, P. et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983 e914 (2018).
Carrion, V. J. et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612 (2019).
Fitzpatrick, C. R. et al. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA 115, E1157–E1165 (2018).
Santos-Medellin, C. et al. Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants 7, 1065–1077 (2021).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Peng, X. & Dorman, K. S. AmpliCI: a high-resolution model-based approach for denoising Illumina amplicon data. Bioinformatics 36, 5151–5158 (2021).
Zhang, P., Spaepen, S., Bai, Y., Hacquard, S. & Garrido-Oter, R. Rbec: a tool for analysis of amplicon sequencing data from synthetic microbial communities. ISME Commun. 1, 73 (2021).
Garrido-Oter, R. et al. Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24, 155–167 e155 (2018).
Karasov, T. L. et al. Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24, 168–179 e164 (2018).
Chiniquy, D. et al. Microbial community field surveys reveal abundant Pseudomonas population in sorghum rhizosphere composed of many closely related phylotypes. Front. Microbiol. 12, 598180 (2021).
Macho, A. P., Zumaquero, A., Ortiz-Martin, I. & Beuzon, C. R. Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae–plant interactions. Mol. Plant Pathol. 8, 437–450 (2007).
Ashe, S., Maji, U. J., Sen, R., Mohanty, S. & Maiti, N. K. Specific oligonucleotide primers for detection of endoglucanase positive Bacillus subtilis by PCR. 3 Biotech 4, 461–465 (2014).
Kebschull, J. M. & Zador, A. M. Cellular barcoding: lineage tracing, screening and beyond. Nat. Methods 15, 871–879 (2018).
Levy, S. F. et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519, 181–186 (2015).
Walsh, C. & Cepko, C. L. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440 (1992).
Lu, R., Neff, N. F., Quake, S. R. & Weissman, I. L. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 29, 928–933 (2011).
Bhang, H. E. et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015).
Abel, S. et al. Sequence tag-based analysis of microbial population dynamics. Nat. Methods 12, 223–226 (2015).
Zhang, T. et al. Deciphering the landscape of host barriers to Listeria monocytogenes infection. Proc. Natl Acad. Sci. USA 114, 6334–6339 (2017).
Wincott, C. J. et al. Cellular barcoding of protozoan pathogens reveals the within-host population dynamics of Toxoplasma gondii host colonization. Cell Rep. Methods 2, 100274 (2022).
Cole, B. J. et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 15, e2002860 (2017).
Luneau, J. S. et al. Genome-wide identification of fitness determinants in the Xanthomonas campestris bacterial pathogen during early stages of plant infection. New Phytol. 236, 235–248 (2022).
Wetmore, K. M. et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6, e00306–e00315 (2015).
Shalev, O. et al. Commensal Pseudomonas strains facilitate protective response against pathogens in the host plant. Nat. Ecol. Evol. 6, 383–396 (2022).
Yu, K. et al. Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 29, 3913–3920 e3914 (2019).
Peters, J. E. & Craig, N. L. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2, 806–814 (2001).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Mitra, R., McKenzie, G. J., Yi, L., Lee, C. A. & Craig, N. L. Characterization of the TnsD-attTn7 complex that promotes site-specific insertion of Tn7. Mob. DNA 1, 18 (2010).
Choi, K. H. & Schweizer, H. P. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161 (2006).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6, e16765 (2011).
Silva-Rocha, R. et al. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675 (2013).
Miller, V. L. & Mekalanos, J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583 (1988).
Cheng, T. et al. Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 16, 138–149 (2016).
Banchi, E. et al. PLANiTS: a curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, baz155 (2020).
Silverman, J. D. et al. Measuring and mitigating PCR bias in microbiota datasets. PLoS Comput. Biol. 17, e1009113 (2021).
Vetrovsky, T. & Baldrian, P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 8, e57923 (2013).
Dehio, C. & Meyer, M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540 (1997).
Wang, P. et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Fact. 14, 11 (2015).
Wippel, K. et al. Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota. Nat. Microbiol. 6, 1150–1162 (2021).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Wei, Q. et al. Crystal structure and function of PqqF protein in the pyrroloquinoline quinone biosynthetic pathway. J. Biol. Chem. 291, 15575–15587 (2016).
Fender, J. E. et al. Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl. Environ. Microbiol. 78, 6225–6235 (2012).
Kremer, J. M. et al. Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nat. Protoc. 16, 2450–2470 (2021).
Geddes, B. A., Mendoza-Suarez, M. A. & Poole, P. S. A Bacterial Expression Vector Archive (BEVA) for flexible modular assembly of golden gate-compatible vectors. Front. Microbiol. 9, 3345 (2018).
Iverson, S. V., Haddock, T. L., Beal, J. & Densmore, D. M. CIDAR MoClo: improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth. Biol. 5, 99–103 (2016).
Louca, S., Doebeli, M. & Parfrey, L. W. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6, 41 (2018).
Tkacz, A., Hortala, M. & Poole, P. S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 110 (2018).
Guo, X. X. et al. Host-associated quantitative abundance profiling reveals the microbial load variation of root microbiome. Plant Commun. 1, 1000003 (2020).
Lundberg, D. S. et al. Host-associated microbe PCR (hamPCR) enables convenient measurement of both microbial load and community composition. eLife 10, e66186 (2021).
Sokolovskaya, O. M., Shelton, A. N. & Taga, M. E. Sharing vitamins: cobamides unveil microbial interactions. Science 369, eaba0165 (2020).
Hantke, K. & Friz, S. The TonB-dependent uptake of pyrroloquinoline-quinone (PQQ) and secretion of gluconate by Escherichia coli K-12. Mol. Microbiol. 118, 417–425 (2022).
Venkataram, S., Kuo, H. Y., Hom, E. F. Y. & Kryazhimskiy, S. Mutualism-enhancing mutations dominate early adaptation in a two-species microbial community. Nat. Ecol. Evol. 7, 143–154 (2023).
House, B. L., Mortimer, M. W. & Kahn, M. L. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70, 2806–2815 (2004).
Choi, K. H. et al. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74, 1064–1075 (2008).
Kvitko, B. H. et al. A simple method for construction of pir+ enterobacterial hosts for maintenance of R6K replicon plasmids. BMC Res. Notes 5, 157 (2012).
Bierman, M. et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992).
Choi, K. H. et al. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448 (2005).
Werner, S., Engler, C., Weber, E., Gruetzner, R. & Marillonnet, S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3, 38–43 (2012).
Ma, K. W., Ordon, J. & Schulze-Lefert, P. Gnotobiotic plant systems for reconstitution and functional studies of the root microbiota. Curr. Protoc. 2, e362 (2022).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012).
Getzke, F. et al. Cofunctioning of bacterial exometabolites drives root microbiota establishment. Proc. Natl Acad. Sci. USA 120, e2221508120 (2023).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Villanueva, R. A. M. & Chen, Z. J. ggplot2: elegant graphics for data analysis, 2nd edition. Meas. Interdiscip. Res. Prespect. 17, 160–167 (2019).
Acknowledgements
We thank D. Becker and E. Logemann for technical support. We acknowledge P. Poole (University of Oxford, UK) for providing the pLM449 plasmid containing the tac promoter and F. Getzke (Max Planck Institute for Plant Breeding Research, Germany) for providing root and peat matrix samples containing fungal DNA. We thank N. Donnelly, J. Stuttmann and R. Berendsen for reading and editing the paper. Funding was provided by the Max Planck Society and the German Research Foundation (DFG) under the German Excellence Strategy, EXC number 2048/1 project 390686111 for R.G.-O. and P.S.-L., and SPP 2125 DECRyPT for K.-W.M. and P.S.-L. J.T. was supported by the Alexander von Humboldt Foundation.
Funding
Open access funding provided by Max Planck Society.
Author information
Authors and Affiliations
Contributions
P.S.-L., J.O., K.-W.M., R.G.-O. and B.H. conceptualized the methodology. J.O. and J.T. designed the experiments. R.T.N. cloned coding sequences for fluorophores. J.O. and J.T. generated MoBacTag plasmids. J.O. assembled barcode DNA tags and tested their diversity. J.T. tested spike-in normalization and amplification biases by multiple MoBacTag-tagged strains. J.O. performed microscopy and the proof-of-principle experiment. J.O., J.T. and P.Z. analysed the data. J.O. and J.T. produced the figures. J.O. and P.S.-L. wrote the paper with contributions from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks David Johnston-Monje, Derek Lundberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Conservation of the Tn7 binding site in the At-SPHERE culture collection.
Tn7 transposase binding site within the last coding 36 nts of the glucosamine-6-phosphate synthetase (glmS) gene, displayed as a DNA sequence logo for all At-SPHERE genome drafts. The letter size indicates the nucleotide abundance at each position. The encoded amino acids are depicted below.
Extended Data Fig. 2 Plasmid map of an N-acetyltransferase aac and GFP-encoding pBCC recipient plasmid.
Plasmid map, created with SnapGene, indicating coding sequences (yellow), promoter sequences (green), terminator sequences (grey), origin of transfer and replication (dark blue), protein binding sites (light blue), primer binding sites (purple) and SmaI restriction sites.
Extended Data Fig. 3 Extended validation of the MoBacTag for spike normalization and barcode DNA tag-specific amplification efficiencies.
a,b, Linear correlation of normalized spike-specific read counts with spike concentrations obtained from amplicon sequencing of root (n = 15) and peat (n = 15) matrix samples using fungal ITS- (ITS1/ITS2; a) and barcode-(JT259/ JT262; b) specific primers. c,d, Amplification efficiencies of different barcode DNA tags determined by qPCR using dilution series of genomic DNA from MoBacTag-labeled wild-type WCS358, WCS358:pqqF and WCS358:cyoB using barcode-specific (c), plant ITS-specific (d) and inter-gene chromosome-specific primers. R values indicate Pearson correlation coefficients and p-values indicate significance by a two-sided t-test.
Extended Data Fig. 4 Chromosome integrity and expression of fluorescent tags in taxonomically distinct bacteria.
a, Alignments of PacBio genome assemblies from the unlabeled and MoBacTag-labeled Rhizobiales LjR239 isolated from Lotus japonicus roots. MoBacTag insertion site is indicated by grey lines. b, Bacterial growth of MoBacTag-labeled or unlabeled Rhodanobacter R179 in liquid rich (0.5 TSB) medium in monocultures indicated by absorbance (λ = 600 nm). c, Expression of fluorescent proteins from chromosomally-integrated MoBacTags. Expression of MoBacTag-encoded fluorescent protein was detected using live confocal laser scanning microscopy in liquid medium (observed two times independently) and on A. thaliana roots (observed three times independently). Representative images are shown. Xcv: Xanthomonas campestris pv. vesicatoria. Scale: 20 µm.
Extended Data Fig. 5 The influence of MoBacTag on root microbiota establishment.
a-c, Coordination of the spike-normalized reads from a 15-member synthetic community with wild-type or MoBacTag-labeled P. capeferrum WCS358 in the A. thaliana root (n = 24) and peat matrix (n = 10) compartment (a), upon in silico depletion of barcode reads (b) and additional in silico depletion of WCS358 16S rRNA reads (c). Shapes represent the compartment and colors represent WCS358 derivatives. n values indicate biological samples collected from three independent replicates. Ellipses correspond to Gaussian distributions fitted to each cluster (95% confidence interval). P values indicate statistical significance determined using a permutational analysis of variance (PERMANOVA) test between communities including the labeled or unlabeled WCS358 derivate (permutation = 999, P < 0.05). d,e, The normalized abundance of individual strains in the root (n = 24) (d) and peat matrix (n = 10) (e) compartment upon co-inoculation with the labeled (WCS358:BC) or unlabeled (WCS358) strain. n values indicate biological samples collected from three independent replicates. P-values indicate statistical significance determined using two-sided Dunn’s test. The box plots center on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the 1.5x interquartile range.
Extended Data Fig. 6 The influence of a MoBacTag on selected SynCom members in independent experiments.
The normalized abundance of strains that had significantly different abundances after co-inoculation with the labeled (WCS358:BC) or unlabeled (WCS358) strain when all replicates are analyzed individually (Extended Data Fig. 5d, e) for each independent experiment. n values indicate biological samples. The box plots center on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the 1.5x interquartile range.
Extended Data Fig. 7 Chemical complementation of WCS358:pqqF mutant by extracellular PQQ.
Bacterial growth of Pseudomonas capeferrum wild-type WCS358 and the WCS358:pqqF mutant in unbuffered modified XVM2 minimal medium supplemented with 110 mM glucose and 3 µM PQQ in monocultures indicated by absorbance (λ = 600 nm).
Extended Data Fig. 8 Detection of barcode DNA tag-specific reads from MoBacTag-labeled Rhizobium R13C and R13D in resident microbial communities of two soil types and a peat substrate.
a, Unconstrained PCoA plot showing differences between bacterial communities in unplanted and planted Cologne agricultural soil (Cas1; black; n = 16, n = 16), soil collected near Golm1 (brown; n = 16, n = 16), Jiffy peat matrix (https://www.jiffygroup.com/; red; n = 16, n = 15), and Arabidopsis roots five weeks after plant cultivation in the respective substrates (n = 16 for each substrate). b-d, Spike-normalized 16S rRNA reads at phylum level of unplanted (b), planted (c) Cas or Golm soils or Jiffy peat matrix, and Arabidopsis Col-0 roots. For this analysis, Rhizobium R13C- and R13D-specific 16S rRNA- and barcode DNA tag-specific reads were depleted in silico. e-g, Normalized barcode-specific read counts of Rhizobium R13C (purple) and R13D (pink) in the indicated unplanted substrates (e), planted substrates (f), and root compartment (g). n values indicate biological samples collected from two independent replicates. The box plots center on the median and extend to the 25th and 75th percentiles, and the whiskers extend to the furthest point within the 1.5x interquartile range.
Supplementary information
Supplementary Information
Supplementary text.
Supplementary Tables
Supplementary Tables 1–6.
Source data
Source Data Fig. 3
Unprocessed DNA gel scan.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ordon, J., Thouin, J., Nakano, R.T. et al. Chromosomal barcodes for simultaneous tracking of near-isogenic bacterial strains in plant microbiota. Nat Microbiol 9, 1117–1129 (2024). https://doi.org/10.1038/s41564-024-01619-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-024-01619-8