Abstract
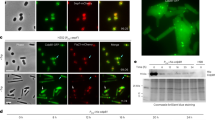
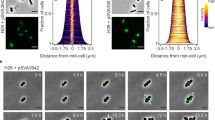
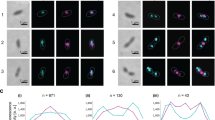
Cell division in all domains of life requires the orchestration of many proteins, but in Archaea most of the machinery remains poorly characterized. Here we investigate the FtsZ-based cell division mechanism in Haloferax volcanii and find proteins containing photosynthetic reaction centre (PRC) barrel domains that play an essential role in archaeal cell division. We rename these proteins cell division protein B 1 (CdpB1) and CdpB2. Depletions and deletions in their respective genes cause severe cell division defects, generating drastically enlarged cells. Fluorescence microscopy of tagged FtsZ1, FtsZ2 and SepF in CdpB1 and CdpB2 mutant strains revealed an unusually disordered divisome that is not organized into a distinct ring-like structure. Biochemical analysis shows that SepF forms a tripartite complex with CdpB1/2 and crystal structures suggest that these two proteins might form filaments, possibly aligning SepF and the FtsZ2 ring during cell division. Overall our results indicate that PRC-domain proteins play essential roles in FtsZ-based cell division in Archaea.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the Source Data. Source data are provided with this paper. Coordinates have been submitted to the Protein Data Bank (PDB) with accession code 8QZO.
References
Mahone, C. R. & Goley, E. D. Bacterial cell division at a glance. J. Cell Sci. 133, jcs237057 (2020).
Andrade, V. & Echard, A. Mechanics and regulation of cytokinetic abscission. Front. Cell Dev. Biol. 10, 1046617 (2022).
Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8, 731–741 (2010).
Ithurbide, S., Gribaldo, S., Albers, S.-V. & Pende, N. Spotlight on FtsZ-based cell division in Archaea. Trends Microbiol. 30, 665–678 (2022).
Makarova, K. S. & Koonin, E. V. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol. Direct 5, 33 (2010).
Liao, Y., Ithurbide, S., Evenhuis, C., Löwe, J. & Duggin, I. G. Cell division in the archaeon Haloferax volcanii relies on two FtsZ proteins with distinct functions in division ring assembly and constriction. Nat. Microbiol. 6, 594–605 (2021).
Pende, N. et al. SepF is the FtsZ-anchor in archaea, with features of an ancestral cell division system. Nat. Commun. https://doi.org/10.1038/s41467-021-23099-8 (2021).
Nußbaum, P., Gerstner, M., Dingethal, M., Erb, C. & Albers, S. V. The archaeal protein SepF is essential for cell division in Haloferax volcanii. Nat. Commun. 12, 3469 (2021).
Anantharaman, V. & Aravind, L. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 3, RESEARCH0061 (2002).
Patro, M., Duggin, I. G., Albers, S.-V. & Ithurbide, S. Influence of plasmids, selection markers and auxotrophic mutations on Haloferax volcanii cell shape plasticity. Front. Microbiol. 14, 1270665 (2023).
Li, Z. et al. Positioning of the motility machinery in halophilic Archaea. mBio 10, e00377-19 (2019).
Ye, H. et al. Crystal structure of the putative adapter protein MTH1859. J. Struct. Biol. 148, 251–256 (2004).
Król, E. et al. Bacillus subtilis SepF binds to the C terminus of FtsZ. PLoS ONE 7, e43293 (2012).
Sogues, A. et al. Essential dynamic interdependence of FtsZ and SepF for Z-ring and septum formation in Corynebacterium glutamicum. Nat. Commun. 11, 1641 (2020).
Elkins, J. G. et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl Acad. Sci. USA 105, 8102–8107 (2008).
Duman, R. et al. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl Acad. Sci. USA 110, E4601–E4610 (2013).
Wenzel, M. et al. Control of septum thickness by the curvature of SepF polymers. Proc. Natl Acad. Sci. USA 118, e2002635118 (2021).
Du, S. & Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 (2017).
Errington, J. & Wu, L. J. in Prokaryotic Cytoskeletons (eds Löwe, J. & Amos, L. A.) 67–101 (Springer, 2017); https://doi.org/10.1007/978-3-319-53047-5_3
Abdul-Halim, M. F. et al. Lipid anchoring of archaeosortase substrates and mid-cell growth in Haloarchaea. mBio 11, 863746 (2020).
Blanch Jover, A. & Dekker, C. The archaeal Cdv cell division system. Trends Microbiol. https://doi.org/10.1016/j.tim.2022.12.006 (2023).
Moriscot, C. et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-Like CdvB. PLoS ONE 6, e21921 (2011).
Samson, R. Y. et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol. Cell 41, 186–196 (2011).
Zhao, S. et al. Widespread PRC barrel proteins are necessary for haloarchaeal cell division. Nat Microbiol. https://doi.org/10.1038/s41564-024-01615-y (2024).
Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 (1951).
Allers, T., Ngo, H.-P., Mevarech, M. & Lloyd, R. G. Development of additional selectable markers for the halophilic Archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 (2004).
de Silva, R. T. et al. Improved growth and morphological plasticity of Haloferax volcanii. Microbiology 167, 001012 (2021).
Watson, J. F. & García-Nafría, J. In vivo DNA assembly using common laboratory bacteria: a re-emerging tool to simplify molecular cloning. J. Biol. Chem. 294, 15271–15281 (2019).
Braun, F. et al. Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. MicrobiologyOpen 8, e00829 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
Lau, Y.-T. K. et al. Discovery and engineering of enhanced SUMO protease enzymes. J. Biol. Chem. 293, 13224–13233 (2018).
Gorrec, F. & Löwe, J. Automated protocols for macromolecular crystallization at the MRC Laboratory of Molecular Biology. J. Vis. Exp. https://doi.org/10.3791/55790 (2018).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
Vonrhein, C. et al. Advances in automated data analysis and processing within autoPROC, combined with improved characterisation, mitigation and visualisation of the anisotropy of diffraction limits using STARANISO. Acta Crystallogr. A 74, a360–a360 (2018).
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Schrödinger, L. & DeLano, W. The PyMOL Molecular Graphics System (Delano Scientific, 2020).
Albers, S. V., Konings, W. N. & Driessen, A. J. in Methods in Microbiology: Extremophiles, Vol. 35 (eds Rainey, F. A. & Oren, A.) 161–171 (Academic Press, 2006).
Scheres, S. H. W. A Bayesian view on Cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Johnson, L. S., Eddy, S. R. & Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinf. 11, 431 (2010).
Garcia, P. S., Gribaldo, S. & Borrel, G. Diversity and evolution of methane-related pathways in Archaea. Annu. Rev. Microbiol. 76, 727–755 (2022).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v.4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Acknowledgements
We thank X. Ye (ISTA) for providing the His–SUMO expression plasmid pSVA13429. pCDB302 was a gift from C. Bahl (Addgene plasmid number 113673; http://n2t.net/addgene:113673; RRID Addgene_113673). We thank B. Ahsan, G. Sharov, G. Cannone and S. Chen from the Medical Research Council (MRC) LMB Electron Microscopy Facility for help and support. We thank Scientific Computing at the MRC LMB for their support. We thank L. Trübestein and N. Krasnici of the protein service unit of the ISTA Lab Support Facility for help with the SEC coupled with multi-angle light scattering experiments. We thank D. Grohmann and R. Reichelt from the Archaea Centre at the University of Regensburg for providing the P. furiosus cell material. P.N. and S.-V.A. were supported by a Momentum grant from the Volkswagen (VW) Foundation (grant number 94933). D.K.-C. and D.B. were supported by the VW Stiftung ‘Life?’ programme (to J.L.; grant number Az 96727) and by the MRC, as part of UK Research and Innovation (UKRI), MRC file reference number U105184326 (to J.L.). N.T. and S.G. acknowledge support from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant number ANR-10-LABX-62-IBEID), and the computational and storage services (Maestro cluster) provided by the IT department at Institut Pasteur. M.K. and M.L. were supported by the Austrian Science Fund (FWF) Stand-Alone P34607. For the purpose of open access, the MRC Laboratory of Molecular Biology has applied a CC BY public copyright licence to any author accepted manuscript version arising.
Author information
Authors and Affiliations
Contributions
P.N. conceived the project and performed all experiments not otherwise mentioned. C.v.d.D. purified the proteins from A. fulgidus and performed SEC experiments. M.K. expressed and purified proteins from H. volcanii and performed SEC coupled with multi-angle light scattering experiments, pelleting assays and mass photometry. D.K.-C. and D.B. solved the crystal structure. D.K.-C., A.Y. and J.L. performed cryo-EM. N.T. performed phylogenetic analysis. M.T. isolated P. furiosus lipids. P.N. and J.L. prepared figures. P.N. wrote the draft of the article. S.G., M.L., J.L. and S.-V.A. reviewed drafts of the article, supervised the work and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Daniela Barilla, William Margolin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Tables 1–6.
Supplementary Video 1
Time lapse of H. volcanii expressing CdpB1–mNeonGreen.
Supplementary Video 2
Time lapse of H. volcanii expressing CdpB2–mNeonGreen.
Supplementary Video 3
Time lapse of H. volcanii expressing CdpB3–mNeonGreen.
Supplementary Video 4
Time lapse of H. volcanii during CdpB1 depletion.
Supplementary Video 5
Time lapse of H. volcanii wild type.
Supplementary Data 1
Source data for supplementary figures.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nußbaum, P., Kureisaite-Ciziene, D., Bellini, D. et al. Proteins containing photosynthetic reaction centre domains modulate FtsZ-based archaeal cell division. Nat Microbiol 9, 698–711 (2024). https://doi.org/10.1038/s41564-024-01600-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-024-01600-5
This article is cited by
-
Finding pieces in the archaeal cell division puzzle
Nature Microbiology (2024)